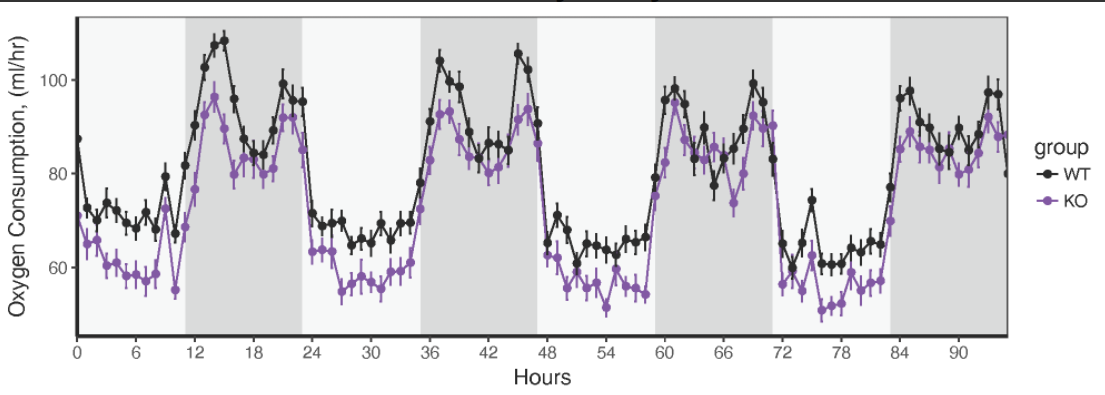
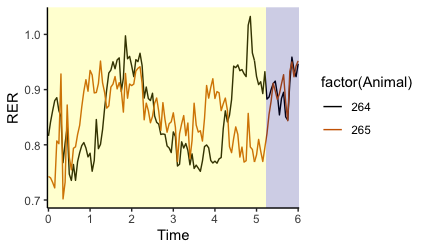
I am working with time-series data, where at every timepoint, I have many measurements for each of up to 16 subjects, spanning multiple days. I currently have the data organized as tidily as I can - here is a snippet.
> df
# A tibble: 23,844 x 40
DateTime Time exp_day Photoperiod ZT Sex Group Treatment Cohort Animal
<dttm> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr> <dbl> <dbl>
1 2021-10-18 12:47:00 0 1 1 6 M mHFD chABC 1 264
2 2021-10-18 12:50:00 0.05 1 1 6 M mHFD chABC 1 264
3 2021-10-18 12:53:00 0.1 1 1 6 M mHFD chABC 1 264
4 2021-10-18 12:56:00 0.15 1 1 6 M mHFD chABC 1 264
5 2021-10-18 12:59:00 0.2 1 1 6 M mHFD chABC 1 264
6 2021-10-18 13:02:00 0.25 1 1 7 M mHFD chABC 1 264
7 2021-10-18 13:05:00 0.3 1 1 7 M mHFD chABC 1 264
8 2021-10-18 13:08:00 0.35 1 1 7 M mHFD chABC 1 264
9 2021-10-18 13:11:00 0.4 1 1 7 M mHFD chABC 1 264
10 2021-10-18 13:14:00 0.45 1 1 7 M mHFD chABC 1 264
... with 23,834 more rows, and 30 more variables
The DateTimes are duplicated for each animal - so while I have 23,844 total observations, there are 1987 for each subject:
> n_distinct(df$DateTime)
[1] 1987
> n_distinct(df$Animal)
[1] 12
What I would like to do is plot time on the x axis, and variables on the y axis, but have the light cycle (light vs. dark) plotted as shaded or light bars behind the data, as shown below.