I'm using FactoMineR's PCA function to compute the PCA of my dataset.
I know I can get the eigenvalues with get_eigenvalue(res.pca) but how do I get the eigenvectors?
CodePudding user response:
In Principal Components Analysis, you're decomposing the n-by-k variable matrix into three parts - U, D and V. U are the left singular vectors that represent the rows of the decomposed matrix. However, the U matrix is not itself the principal components. All of the columns of U are mutually orthogonal (as you would expect), but they also all have the same variance. The principal components are U scaled by the square root of the eigenvalues. Here's an example:
library(FactoMineR)
data(decathlon)
dat <- decathlon[,1:10]
p <- PCA(dat)
#> Warning: ggrepel: 1 unlabeled data points (too many overlaps). Consider
#> increasing max.overlaps
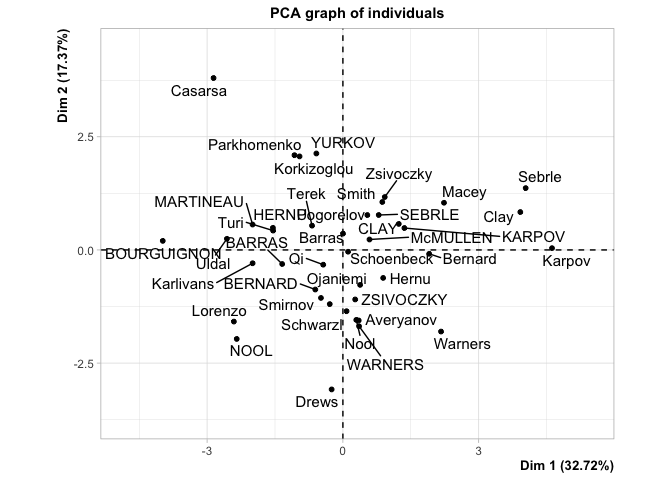
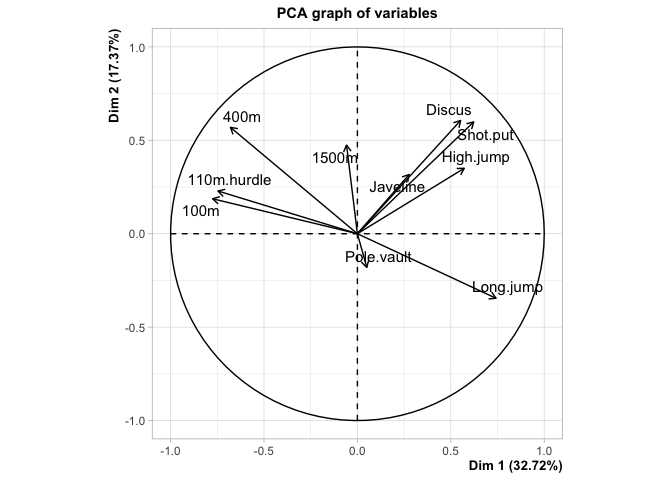
comp1 <- predict(p, newdata=dat)$coord[,1:3]
U <- p$svd$U
e <- p$eig[,"eigenvalue"]
comp2 <- U[,1:3] %*% diag(sqrt(e[1:3]))
head(comp1[,1:3])
#> Dim.1 Dim.2 Dim.3
#> SEBRLE 0.7916277 0.7716112 0.8268412
#> CLAY 1.2349906 0.5745781 2.1412470
#> KARPOV 1.3582149 0.4840209 1.9562580
#> BERNARD -0.6095151 -0.8746285 0.8899407
#> YURKOV -0.5859683 2.1309542 -1.2251568
#> WARNERS 0.3568895 -1.6849567 0.7665531
head(comp2[,1:3])
#> [,1] [,2] [,3]
#> [1,] 0.7916277 0.7716112 0.8268412
#> [2,] 1.2349906 0.5745781 2.1412470
#> [3,] 1.3582149 0.4840209 1.9562580
#> [4,] -0.6095151 -0.8746285 0.8899407
#> [5,] -0.5859683 2.1309542 -1.2251568
#> [6,] 0.3568895 -1.6849567 0.7665531
Created on 2022-02-16 by the reprex package (v2.0.1)
