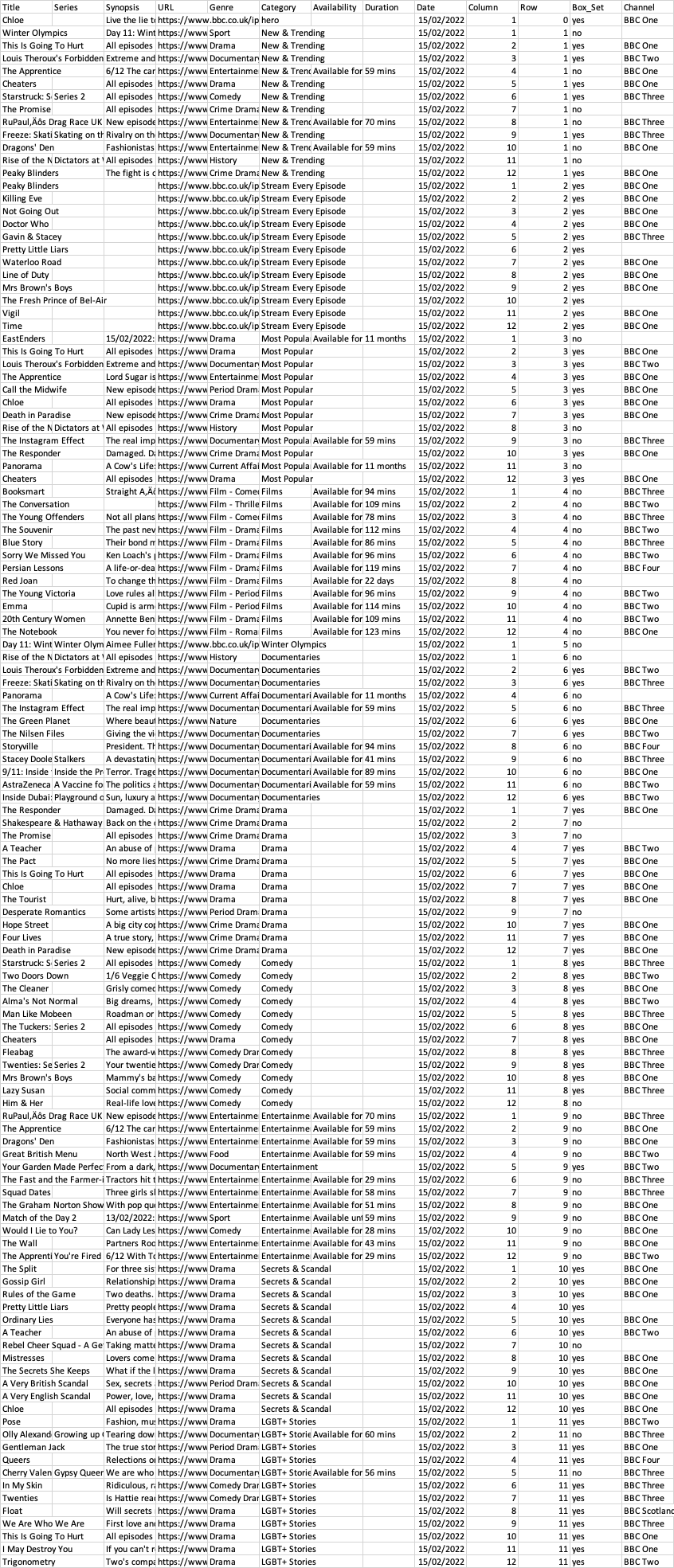
I have a large and complex dataset (a bit too complex to share here, and probably not necessary to share the whole thing) but here's an example of what it looks like. This is just one day and the full sample spans hundreds of days:
What I want to do, is to devise a way to count variation of the Genre within each Row. To put it more simply (I hope): each Row has 12 Columns and I want to measure the variation of Genre across those 12 Columns (it's the BBC iPlayer, which many of you might be familiar with). E.g. If a Row is comprised of 4 "sport", 4 "drama", and 4 "documentary", there would be a distinct count of 3 genres.
I'm thinking that a simple distinct count would be a good way to measure variation within each row (the more distinct the count, the higher the variation) but it's not a very nuanced approach. I.e. if a row is comprised of 11 "sport" and 1 "documentary" it's a distinct count of 2. If it's comprised of 6 "sport" and 6 "documentary" it's still a distinct count of 2 - so distinct count doesn't really help in that sense.
I guess I'm asking for advice on two things here:
- Firstly, what would be the most appropriate way to measure variation
of
Genrewithin eachRow - Secondly, how would I go about doing that! I.e. what code / packages would I need?
I hope that's all clear, but if not, I'd be happy to elaborate on anything. It's perhaps worth noting (as I mentioned above) that I want to determine variation on a specific date, and the sample data shared here is just one date (but I have hundreds).
Thanks in advance :)
*** Update ***
Thanks for the comments below - especially about sharing a snapshot of the real data (which you'll find below). My apologies - I'm a bit of a novice in this area and not really familiar with the proper conventions!
Here's a sample of the data - I hope it's right and I hope it helps:
structure(list(Row = c(0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 3L), Genre = c("", "Sport", "Drama", "Documentary",
"Entertainment", "Drama", "Comedy", "Crime Drama", "Entertainment",
"Documentary", "Entertainment", "History", "Crime Drama", "",
"", "", "", "", "", "", "", "", "", "", "", "Drama", "Drama",
"Documentary", "Entertainment", "Period Drama"), Column = c(1L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L)), row.names = c(NA,
30L), class = "data.frame")
CodePudding user response:
First create some reproducible data. All we need is Row and Genre:
set.seed(42)
Row <- rep(1:10, each=10)
Genre <- sample(c("Sport", "Drama", "Documentary", "Entertainment", "History", "Crime Drama", "Period Drama", "Film - Comedy", "Film-Thriller"), 100, replace=TRUE)
example <- data.frame(Row, Genre)
str(example)
# 'data.frame': 100 obs. of 2 variables:
# $ Row : int 1 1 1 1 1 1 1 1 1 1 ...
# $ Genre: chr "Sport" "History" "Sport" "Film-Thriller" ...
Now to get the number of different genres in each row:
Count <- tapply(example$Genre, example$Row, function(x) length(unique(x)))
Count
# 1 2 3 4 5 6 7 8 9 10
# 7 5 6 7 6 8 7 7 7 6
There are 7 genres in row 1 and only 5 in row 2. For more detail:
xtabs(~Genre Row, example)
# Row
# Genre 1 2 3 4 5 6 7 8 9 10
# Crime Drama 0 0 1 1 3 1 1 0 2 1
# Documentary 0 1 1 1 1 0 1 0 1 0
# Drama 1 1 1 3 2 1 2 1 1 0
# Entertainment 2 2 3 1 1 1 1 2 0 0
# Film - Comedy 1 0 3 2 0 1 2 2 0 2
# Film-Thriller 1 3 0 0 0 1 1 1 2 2
# History 1 3 0 1 2 1 2 2 2 1
# Period Drama 1 0 0 1 0 2 0 1 1 2
# Sport 3 0 1 0 1 2 0 1 1 2
CodePudding user response:
Reproducible sample data:
set.seed(42)
sampdata <- transform(
expand.grid(Date = Sys.Date() 0:2, Row=0:3, Column=1:12),
Genre = sample(c("Crime Drama","Documentary","Drama","Entertainment"),
size = 48, replace = TRUE)
)
head(sampdata)
# Date Row Column Genre
# 1 2022-02-18 0 1 Crime Drama
# 2 2022-02-19 0 1 Crime Drama
# 3 2022-02-20 0 1 Crime Drama
# 4 2022-02-18 1 1 Crime Drama
# 5 2022-02-19 1 1 Documentary
# 6 2022-02-20 1 1 Entertainment
nrow(sampdata)
# [1] 144
Using dplyr and tidyr, we can group, summarize, then pivot:
library(dplyr)
# library(tidyr) # pivot_wider
sampdata %>%
group_by(Date, Row) %>%
summarize(
Uniq = n_distinct(Genre),
Var = var(table(Genre))
) %>%
tidyr::pivot_wider(
Date, names_from = Row, values_from = c(Uniq, Var)
) %>%
ungroup()
# # A tibble: 3 x 9
# Date Uniq_0 Uniq_1 Uniq_2 Uniq_3 Var_0 Var_1 Var_2 Var_3
# <date> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
# 1 2022-02-18 2 3 2 2 0 3 18 18
# 2 2022-02-19 3 3 1 3 3 3 NA 3
# 3 2022-02-20 2 3 3 3 18 3 3 3
Two things: Uniq_# is per-Row counts of distinct Genre values, and Var_# are the variance of the counts. For instance, in your example, two genres with counts 6 and 6 will have a variance of 0, but counts of 11 and 1 will have a variance of 50 (var(c(11,1))), indicating more variation for that Date/Row combination.
Because we use group_by, if you have even more grouping variables, it is straight-forward to extend this, both in the grouping and in what aggregation we can do in addition to n_distinct(.) and var(.).
BTW: depending on your other calculations, analysis, and reporting/plotting, it might be useful to keep this in the long format, removing the pivot_wider.
sampdata %>%
group_by(Date, Row) %>%
summarize(
Uniq = n_distinct(Genre),
Var = var(table(Genre))
) %>%
ungroup()
# # A tibble: 12 x 4
# Date Row Uniq Var
# <date> <int> <int> <dbl>
# 1 2022-02-18 0 2 0
# 2 2022-02-18 1 3 3
# 3 2022-02-18 2 2 18
# 4 2022-02-18 3 2 18
# 5 2022-02-19 0 3 3
# 6 2022-02-19 1 3 3
# 7 2022-02-19 2 1 NA
# 8 2022-02-19 3 3 3
# 9 2022-02-20 0 2 18
# 10 2022-02-20 1 3 3
# 11 2022-02-20 2 3 3
# 12 2022-02-20 3 3 3
Good examples of when to keep it long include further aggregation by Date/Row and plotting with ggplot2 (which really rewards long-data.