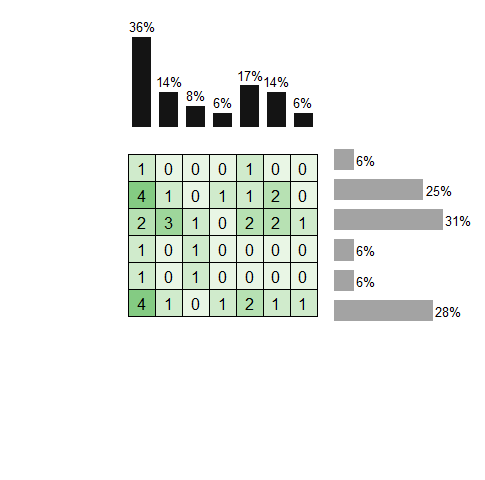
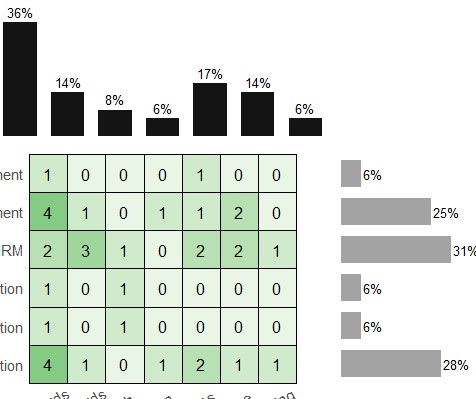
When using geom_tile() to make a ggplot object with theme_void() there are blank areas at top and bottom of plot, and when using patchwork to add pieces these cause a mis-alignment of right=side marginal bars (as in screenshot attached).
Please is there a way to eliminate the blank spaces? I tried a custom theme (code further below) but it didnt help (and sometimes put space on left/right sides also)
library(tidyverse)
library(grid)
library(patchwork)
long <- dget('long.txt')
# Summarise data for marginal plots
in_y_df <- long %>%
group_by(in_y) %>%
summarise(value = sum(value)) %>%
mutate(value = value / sum(value)) %>%
mutate(str = paste0( round(value * 100, digits=0), '%' ))
out_x_df <- long %>%
group_by(out_x) %>%
summarise(value = sum(value)) %>%
mutate(value = value / sum(value)) %>%
mutate(str = paste0( round(value * 100, digits=0), '%' ))
# Marginal plots
py <- ggplot(in_y_df, aes(value, in_y))
geom_col(width = .7, fill = 'gray64')
geom_text(aes(label = str), hjust = -0.1, size = 10 / .pt)
scale_x_continuous(expand = expansion(mult = c(.0, .25)))
theme_void()
theme(plot.margin = unit(c(0,0,0,0), "mm"))
px <- ggplot(out_x_df, aes(out_x, value))
geom_col(width = .7, fill = 'gray8')
geom_text(aes(label = str), vjust = -0.5, size = 10 / .pt)
scale_y_continuous(expand = expansion(mult = c(.0, .25)))
theme_void()
theme(plot.margin = unit(c(0,0,0,0), "mm"))
h2 <- ggplot(long, aes(out_x, in_y, fill = value))
geom_tile(color = 'black', size = 0.2)
coord_equal()
geom_text(aes(label = value), size = 12 / .pt)
scale_fill_gradient(low = "#E9F6E5", high = "#84CB83")
theme_void()
theme(legend.position = 0)
labs(x = NULL, y = NULL, fill = NULL)
heatmap_minus_axisText <- h2
# Version with axis labels not in heatplot
yLabelsPlot <- plot_spacer()
xLabelsPlot <- plot_spacer()
squarePlot <- plot_spacer() px plot_spacer()
yLabelsPlot heatmap_minus_axisText py
plot_spacer() xLabelsPlot plot_spacer()
plot_layout(ncol = 3, widths = c(1, 2, 1.4), heights = c(1.2, 2, 1.3))
squarePlot
The axis labels (not yet made) will be done separately because when included in heatmap object they cause top-bars to misalign after stitching with patchwork, as shown in 2nd picture.
heatPlot <- plot_spacer() px plot_spacer()
plot_spacer() ph py
plot_layout(ncol = 3, widths = c(1, 2, 0.8), heights = c(1.2, 2))
Here is the file for dget() to make fake dataframe:
structure(list(in_y = structure(c(4L, 3L, 6L, 2L, 1L, 5L, 4L,
3L, 6L, 2L, 1L, 5L, 4L, 3L, 6L, 2L, 1L, 5L, 4L, 3L, 6L, 2L, 1L,
5L, 4L, 3L, 6L, 2L, 1L, 5L, 4L, 3L, 6L, 2L, 1L, 5L, 4L, 3L, 6L,
2L, 1L, 5L), .Label = c("R5BnqKkCI3iHHfBDOd3D", "pJoPxfMkRaLbl2ESDD3s",
"8vXGhUVjAubzkubDQft5", "rtUdC7ZmBc8GfiyyJ9DF", "teSB1z4wpHzLLmqwLR4t",
"55G18mbzeqajtzO7WLLM"), class = "factor"), out_x = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 4L, 4L, 4L, 4L, 4L, 4L, 6L, 6L, 6L, 6L, 6L,
6L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 5L, 5L, 5L,
5L, 5L, 5L, 7L, 7L, 7L, 7L, 7L, 7L), .Label = c("MCbgrg3f1oVlszlUUddq",
"yjqUPElzDhsAHKpVPLo6", "j3tZCBrw6qY9X2pYDc9Y", "iW2TVkR3NawpBC8p59F2",
"L1Ai51LaakWu0b47zypc", "HjkPWE5auDZ0dOH3G1L1", "WEfxf3XV0CMSWRRLHheY"
), class = "factor"), value = c(2L, 1L, 1L, 1L, 4L, 4L, 0L, 0L,
0L, 0L, 1L, 1L, 2L, 0L, 0L, 0L, 1L, 2L, 1L, 1L, 0L, 1L, 0L, 0L,
3L, 0L, 0L, 0L, 1L, 1L, 2L, 0L, 1L, 0L, 2L, 1L, 1L, 0L, 0L, 0L,
1L, 0L)), row.names = c(NA, -42L), class = "data.frame")
custom theme
heatmap_theme <- theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
axis.ticks.length = unit(0, "pt"),
legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
plot.margin = unit(c(0,0,0,0), "mm"),
panel.spacing = unit(c(0,0,0,0), "mm")
)
CodePudding user response:
You could try to specify axis limits and suppress automatic axis expansion. Setting axis limits is more straightforward for factors: if needed, convert numeric tile coordinates to character (as.character) or factor (as.factor). Also remove the legend.
Example of stripped heatmap:
data.frame(
x = c('col_1','col_2','col_1','col_2'),
y = c('row_1','row_1','row_2','row_2'),
value = runif(4)
) %>%
ggplot()
geom_tile(aes(x, y, fill = value))
scale_x_discrete(limits = c('col_1','col_2'), expand = c(0,0))
scale_y_discrete(limits = c('row_1','row_2'), expand = c(0,0))
scale_fill_continuous(guide = 'none')
theme_void()
edit: for composite charts, there's also cowplot