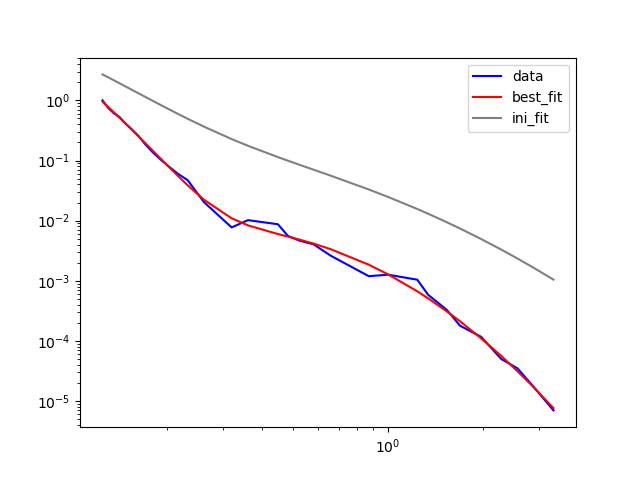
I have been spending FAR too much time trying to figure this out - so time to seek help. I am attempting to use lmfit to fit two lognormals (a and c) as well as the sum of these two lognormals (a c) to a size distribution. Mode a centers around x=0.2, y=1, mode c centers around x=1.2, y=<<<1. There are numerous size distributions (>200) which are all slightly different and are passed in to the following code from an outside loop. For this example, I have provided a real life distribution and have not included the loop. Hopefully my code is sufficiently annotated to allow understanding of what I am trying to achieve.
I must be missing some fundamental understanding of lmfit (spoiler alert - I'm not great at Maths either) as I have 2 problems:
- the fits (a, c and a c) do not accurately represent the data. Note how the fit (red solid line) diverts away from the data (blue solid line). I assume this is something to do with the initial guess parameters. I have tried LOTS and have been unable to get a good fit.
- re-running the model with "new" best fit values (results2, results3) doesn't appear to significantly improve the fit at all. Why?
Example result using provided x and y data:
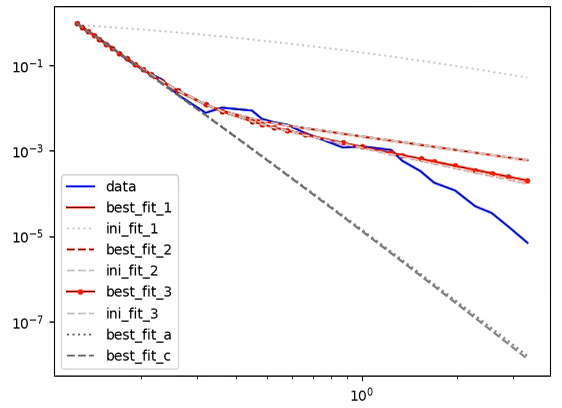
Here is one-I-made-earlier showing the type of fit I am after (produced using the older mpfit module, using different data than provided below and using unique initial best guess parameters (not in a loop). Excuse the legend format, I had to remove certain information):
Any assistance is much appreciated. Here is the code with an example distribution:
from lmfit import models
import matplotlib.pyplot as plt
import numpy as np
# real life data example
y = np.array([1.000000, 0.754712, 0.610303, 0.527856, 0.412125, 0.329689, 0.255756, 0.184424, 0.136819,
0.102316, 0.078763, 0.060896, 0.047118, 0.020297, 0.007714, 0.010202, 0.008710, 0.005579,
0.004644, 0.004043, 0.002618, 0.001194, 0.001263, 0.001043, 0.000584, 0.000330, 0.000179,
0.000117, 0.000050, 0.000035, 0.000017, 0.000007])
x = np.array([0.124980, 0.130042, 0.135712, 0.141490, 0.147659, 0.154711, 0.162421, 0.170855, 0.180262,
0.191324, 0.203064, 0.215738, 0.232411, 0.261810, 0.320252, 0.360761, 0.448802, 0.482528,
0.525526, 0.581518, 0.658988, 0.870114, 1.001815, 1.238899, 1.341285, 1.535134, 1.691963,
1.973359, 2.285620, 2.572177, 2.900414, 3.342739])
# create the joint model using prefixes for each mode
model = (models.LognormalModel(prefix='p1_')
models.LognormalModel(prefix='p2_'))
# add some best guesses for the model parameters
params = model.make_params(p1_center=0.1, p1_sigma=2, p1_amplitude=1,
p2_center=1, p2_sigma=2, p2_amplitude=0.000000000000001)
# bound those best guesses
# params['p1_amplitude'].min = 0.0
# params['p1_amplitude'].max = 1e5
# params['p1_sigma'].min = 1.01
# params['p1_sigma'].max = 5
# params['p1_center'].min = 0.01
# params['p1_center'].max = 1.0
#
# params['p2_amplitude'].min = 0.0
# params['p2_amplitude'].max = 1
# params['p2_sigma'].min = 1.01
# params['p2_sigma'].max = 10
# params['p2_center'].min = 1.0
# params['p2_center'].max = 3
# actually fit the model
result = model.fit(y, params, x=x)
# ====================================
# ================================
# re-run using the best-fit params derived above
params2 = model.make_params(p1_center=result.best_values['p1_center'], p1_sigma=result.best_values['p1_sigma'],
p1_amplitude=result.best_values['p1_amplitude'],
p2_center=result.best_values['p2_center'], p2_sigma=result.best_values['p2_sigma'],
p2_amplitude=result.best_values['p2_amplitude'], )
# re-fit the model
result2 = model.fit(y, params2, x=x)
# ================================
# re-run again using the best-fit params derived above
params3 = model.make_params(p1_center=result2.best_values['p1_center'], p1_sigma=result2.best_values['p1_sigma'],
p1_amplitude=result2.best_values['p1_amplitude'],
p2_center=result2.best_values['p2_center'], p2_sigma=result2.best_values['p2_sigma'],
p2_amplitude=result2.best_values['p2_amplitude'], )
# re-fit the model
result3 = model.fit(y, params3, x=x)
# ================================
# add individual fine and coarse modes using the revised fit parameters
model_a = models.LognormalModel()
params_a = model_a.make_params(center=result3.best_values['p1_center'], sigma=result3.best_values['p1_sigma'],
amplitude=result3.best_values['p1_amplitude'])
result_a = model_a.fit(y, params_a, x=x)
model_c = models.LognormalModel()
params_c = model_c.make_params(center=result3.best_values['p2_center'], sigma=result3.best_values['p2_sigma'],
amplitude=result3.best_values['p2_amplitude'])
result_c = model_c.fit(y, params_c, x=x)
# ====================================
plt.plot(x, y, 'b-', label='data')
plt.plot(x, result.best_fit, 'r-', label='best_fit_1')
plt.plot(x, result.init_fit, 'lightgrey', ls=':', label='ini_fit_1')
plt.plot(x, result2.best_fit, 'r--', label='best_fit_2')
plt.plot(x, result2.init_fit, 'lightgrey', ls='--', label='ini_fit_2')
plt.plot(x, result3.best_fit, 'r.-', label='best_fit_3')
plt.plot(x, result3.init_fit, 'lightgrey', ls='--', label='ini_fit_3')
plt.plot(x, result_a.best_fit, 'grey', ls=':', label='best_fit_a')
plt.plot(x, result_c.best_fit, 'grey', ls='--', label='best_fit_c')
plt.xscale("log")
plt.yscale("log")
plt.legend()
plt.show()
CodePudding user response:
There are three main pieces of advice I can give:
- initial values matter and should not be so far off as to make portions of the model completely insensitive to the parameter values. Your initial model is sort of off by several orders of magnitude.
- always look at the fit result. This is the primary result -- the plot of the fit is a representation of the actual numerical results. Not showing that you printed out the fit report is a good indication that you did not look at the actual result. Really, always look at the results.
- if you are judging the quality of the fit based on a plot of the data and model, use how you choose to plot the data to guide how you fit the data. Specifically in your case, if you are plotting on a log scale, then fit the log of the data to the log of the model: fit in "log space".
Such a fit might look like this:
from lmfit import models, Model
from lmfit.lineshapes import lognormal
import matplotlib.pyplot as plt
import numpy as np
y = np.array([1.000000, 0.754712, 0.610303, 0.527856, 0.412125, 0.329689, 0.255756, 0.184424, 0.136819,
0.102316, 0.078763, 0.060896, 0.047118, 0.020297, 0.007714, 0.010202, 0.008710, 0.005579,
0.004644, 0.004043, 0.002618, 0.001194, 0.001263, 0.001043, 0.000584, 0.000330, 0.000179,
0.000117, 0.000050, 0.000035, 0.000017, 0.000007])
x = np.array([0.124980, 0.130042, 0.135712, 0.141490, 0.147659, 0.154711, 0.162421, 0.170855, 0.180262,
0.191324, 0.203064, 0.215738, 0.232411, 0.261810, 0.320252, 0.360761, 0.448802, 0.482528,
0.525526, 0.581518, 0.658988, 0.870114, 1.001815, 1.238899, 1.341285, 1.535134, 1.691963,
1.973359, 2.285620, 2.572177, 2.900414, 3.342739])
# use a model that is the log of the sum of two log-normal functions
# note to be careful about log(x) for x < 0.
def log_lognormal(x, amp1, cen1, sig1, amp2, cen2, sig2):
comp1 = lognormal(x, amp1, cen1, sig1)
comp2 = lognormal(x, amp2, cen2, sig2)
total = comp1 comp2
total[np.where(total<1.e-99)] = 1.e-99
return np.log(comp1 comp2)
model = Model(log_lognormal)
params = model.make_params(amp1=5.0, cen1=-4, sig1=1,
amp2=0.1, cen2=-1, sig2=1)
# part of making sure that the lognormals are strictly positive
params['amp1'].min = 0
params['amp2'].min = 0
result = model.fit(np.log(y), params, x=x)
print(result.fit_report()) # <-- HERE IS WHERE THE RESULTS ARE!!
# also, make a plot of data and fit
plt.plot(x, y, 'b-', label='data')
plt.plot(x, np.exp(result.best_fit), 'r-', label='best_fit')
plt.plot(x, np.exp(result.init_fit), 'grey', label='ini_fit')
plt.xscale("log")
plt.yscale("log")
plt.legend()
plt.show()
This will print out
[[Model]]
Model(log_lognormal)
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 211
# data points = 32
# variables = 6
chi-square = 0.91190970
reduced chi-square = 0.03507345
Akaike info crit = -101.854407
Bayesian info crit = -93.0599914
[[Variables]]
amp1: 21.3565856 /- 193.951379 (908.16%) (init = 5)
cen1: -4.40637490 /- 3.81299642 (86.53%) (init = -4)
sig1: 0.77286862 /- 0.55925566 (72.36%) (init = 1)
amp2: 0.00401804 /- 7.5833e-04 (18.87%) (init = 0.1)
cen2: -0.74055538 /- 0.13043827 (17.61%) (init = -1)
sig2: 0.64346873 /- 0.04102122 (6.38%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amp1, cen1) = -0.999
C(cen1, sig1) = -0.999
C(amp1, sig1) = 0.997
C(cen2, sig2) = -0.964
C(amp2, cen2) = -0.940
C(amp2, sig2) = 0.849
C(sig1, amp2) = -0.758
C(cen1, amp2) = 0.740
C(amp1, amp2) = -0.726
C(sig1, cen2) = 0.687
C(cen1, cen2) = -0.669
C(amp1, cen2) = 0.655
C(sig1, sig2) = -0.598
C(cen1, sig2) = 0.581
C(amp1, sig2) = -0.567
and generate a plot like