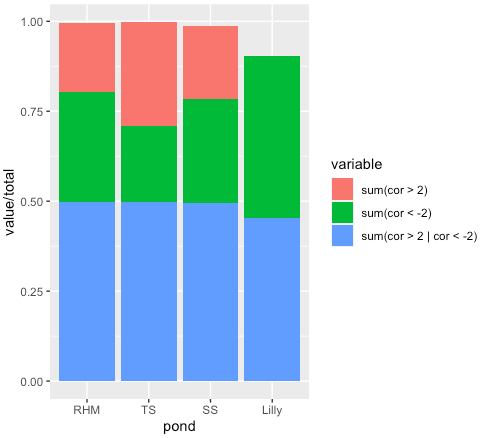
I'm looking to normalize values while retaining their relative frequency. For example, the total counts for one variable is 219, comprised of the values 56, 89, 145. To normalize these data I divided each value by the total then visualized the results as a bar chart, shown below. Why aren't the total values summing to 1.00?
p.perc <- ggplot(bNTI.perc, aes(fill=variable,x=pond,y=value/total))
geom_bar(stat = "identity")
print (p.perc)
Thank you! My data:
> dput(bNTI.perc)
structure(list(pond = structure(c(1L, 2L, 3L, 4L, 1L, 2L, 3L,
4L, 1L, 2L, 3L, 4L), .Label = c("RHM", "TS", "SS", "Lilly"), class = "factor"),
total = c(291, 740, 241, 42, 291, 740, 241, 42, 291, 740,
241, 42), variable = structure(c(1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L), .Label = c("sum(cor > 2)", "sum(cor < -2)",
"sum(cor > 2 | cor < -2)"), class = "factor"), value = c(56L,
213L, 49L, 0L, 89L, 156L, 70L, 19L, 145L, 369L, 119L, 19L
)), row.names = c(NA, -12L), class = "data.frame")
CodePudding user response:
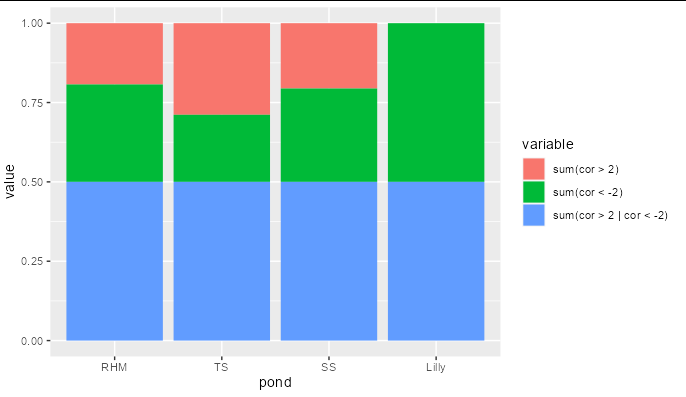
You don't need to do this manually. Use position = 'fill':
ggplot(bNTI.perc, aes(pond, value, fill = variable)) geom_col(position = 'fill')
But the answer to your actual question is that your total column is wrong. There are three values for Lilly (0, 19 and 19), which sums to 38, yet your total for the Lilly group is 42, not 38, so your Lilly bar was only adding up to 38/42 (0.9047619). Similarly, your SS values of 119 70 49 add up to 238, yet your total for SS is 241