I've searched everywhere on the internet but I cannot find an answer to my problem. I have plotted a smooth regression line with confidence intervals with ggplot, but the confidence area is not displayed as a band around the regression line, but only below the line. Is this a graphing error or am I missing the right mathematics as an explanation?
I've used the following code:
library(dplyr); library(ggplot2)
fig <-
df %>%
mutate(
Legend = case_when(
ind == "currentLifetimeRisk" ~ "Lifetime risk",
ind == "currentTenYearRisk" ~ "Ten year risk"
)
) %>%
ggplot(aes(x = age_measurements,
y = values,
fill=Legend))
geom_smooth(
aes(ymin = 0,
ymax = ..y..,),
alpha = 0.8,
formula = y ~ s(x, k = 9, bs="cs"),
color= "black", # kleur van de lijn
stat = "smooth",
method = "gam",
se = TRUE
)
scale_y_continuous(breaks = seq(0, 100, 25),
limits = c(0, 100), expand = c(0,0))
scale_x_continuous(breaks = seq(30, max(fig2_long$age_measurements), 10), expand = c(0,0))
theme(
axis.text =element_text(family = "sans", size=18,colour = "black"),
axis.title = element_text(size=18),
axis.line.x = element_line(size=0.5, linetype="solid", colour="black"),
axis.line.y = element_line(size=0.5, linetype="solid", colour="black"),
panel.grid.major = element_line(colour = "lightgrey"),
panel.background = element_blank(),
)
labs(
x = "Age at risk estimation",
y = "Risk of recurrent events (%)",
)
fig 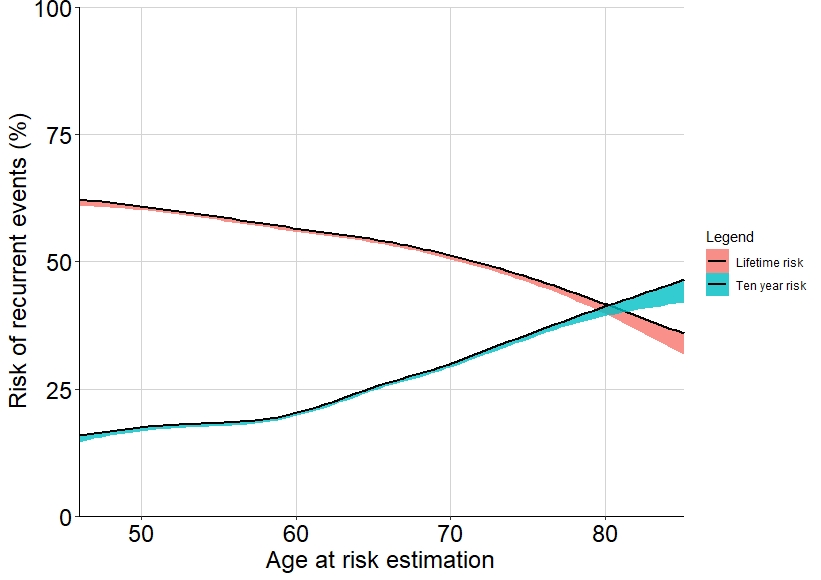
I expected to see the 95% conf interval as a band around the regression line, not only below. Any help is highly appreciated.
CodePudding user response:
The use of aes(ymax=..y..) is forcing the top of the confidence interval to be equal to the fitted y value of the regression line calculated by geom_smooth. If you remove aes(ymin = 0, ymax = ..y..), the correct confidence intervals will be plotted.
..y.., ..ymax.., and ..ymin.. are values calculated internally by geom_smooth in order to plot the regression line and confidence intervals. Setting ymax=..y.. forces geom_smooth to use ..y.. (the fitted y value of the regression) rather than ..ymax.. (the calculated top of the 95% confidence interval) as the top of the 95% confidence interval in the plot, resulting in the issue you're seeing. (I'm actually not sure why ymin=0 isn't forcing the bottom of the 95% confidence interval to be zero.)
Removing the aes mappings (which aren't needed for geom_smooth) results in the expected plotting of the confidence intervals. The example below reproduces the issue you're seeing:
library(tidyverse)
library(patchwork)
p1 = mtcars %>%
ggplot(aes(mpg, hp, fill=factor(vs)))
geom_smooth(
aes(ymin=0, ymax = ..y..,),
alpha = 0.8,
formula = y ~ s(x, k=9, bs="cs"),
color= "black", # kleur van de lijn
stat = "smooth",
method = "gam",
se = TRUE
)
p2 = mtcars %>%
ggplot(aes(mpg, hp, fill=factor(vs)))
geom_smooth(
alpha = 0.8,
formula = y ~ s(x, k=9, bs="cs"),
color= "black", # kleur van de lijn
stat = "smooth",
method = "gam",
se = TRUE
)
p1 p2
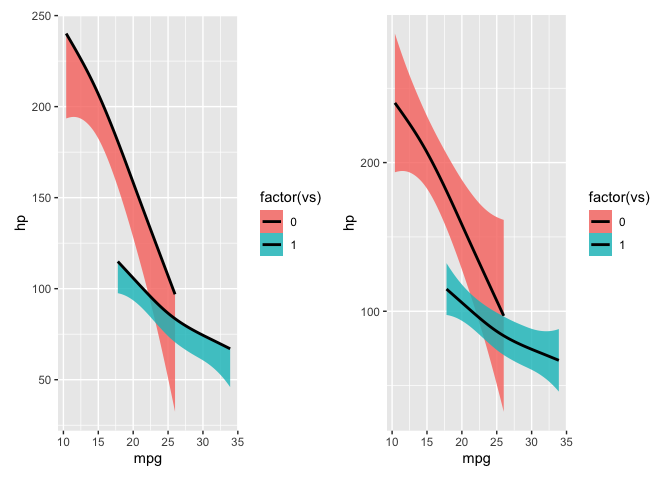
Created on 2022-03-23 by the reprex package (v2.0.1)
