So, I have this scatterplot:
data(infmort, package = "faraway")
summary(infmort)
#install.packages("ggplot2")
library(ggplot2)
#levels(infmort$region)
# Levels are : 1) Africa, 2) Europe, 3) Asia, 4), Americas
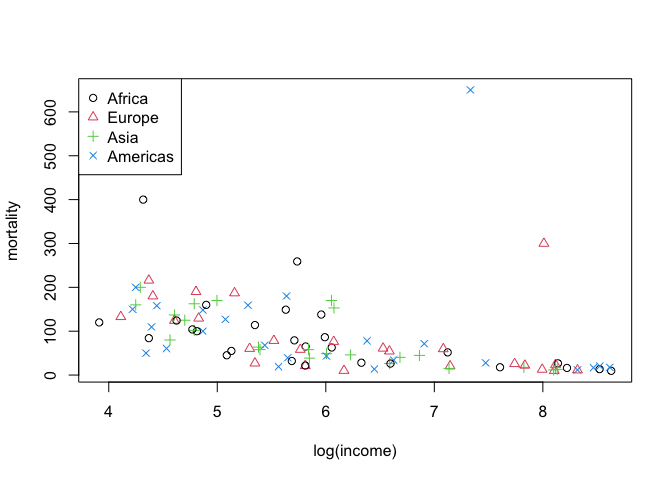
plot(mortality ~ log(income), data = infmort, col = region_col, pch = region_col)
legend("topleft", legend = c("Africa", "Europe", "Asia", "Americas"), col = 1:4, pch = 1:4)
I want to drop the levels of "Africa" and "Europe", so that my scatterplot is only showing the data for Asia and Americas. How do I go about doing this? I am new to R.
Here is what I tried to drop these two categorial variables:
plot(mortality ~ log(income), data=infmort, col = typ_col, pch = typ_col)
legend("topleft", legend = c("Americas", "Asia"), col = 1:2, pch= 1:2)
I know that this is incorrect because it is still including all of the data, I was just wondering how I only graph the data for Americas and Asia.
CodePudding user response:
One potential solution is to subset the dataframe before plotting, e.g.
#install.packages("faraway")
data(infmort, package = "faraway")
summary(infmort)
#> region income mortality oil
#> Africa :34 Min. : 50.0 Min. : 9.60 oil exports : 9
#> Europe :18 1st Qu.: 123.0 1st Qu.: 26.20 no oil exports:96
#> Asia :30 Median : 334.0 Median : 60.60
#> Americas:23 Mean : 998.1 Mean : 89.05
#> 3rd Qu.:1191.0 3rd Qu.:129.40
#> Max. :5596.0 Max. :650.00
#> NA's :4
#install.packages("ggplot2")
library(ggplot2)
#levels(infmort$region)
# Levels are : 1) Africa, 2) Europe, 3) Asia, 4), Americas
region_col <- 1:4
plot(mortality ~ log(income), data = infmort, col = region_col, pch = region_col)
legend("topleft", legend = c("Africa", "Europe", "Asia", "Americas"), col = 1:4, pch = 1:4)
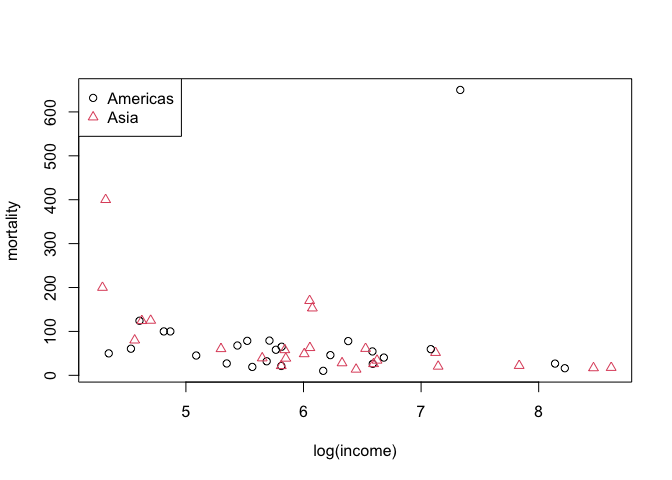
typ_col <- 1:2
plot(mortality ~ log(income),
data=infmort[infmort$region %in% c("Americas", "Asia"),],
col = typ_col, pch = typ_col)
legend("topleft", legend = c("Americas", "Asia"), col = 1:2, pch= 1:2)
Created on 2022-03-30 by the reprex package (v2.0.1)
CodePudding user response:
You can simply filter the data prior to making a plot.
library(dplyr)
infmort <- infmort %>% dplyr::filter(region %in% c("Asia", "America"))
