Few requirements.
Before posting your answer please!!
1) Make sure that your function does not give errors with other data, simulate several similar matrices. (turn off the seed)
2) Make sure your function is faster than mine
3) Make sure that your function works exactly the same as mine, simulate it on different matrices (turn off the seed)
for example
for(i in 1:500){
m <- matrix(sample(c(F,T),30,T),ncol = 3) ; colnames(m) <- paste0("x",1:ncol(m))
res <- c(my_fun(m),your_function(m))
print(res)
if(sum(res)==1) break
}
m
At the moment I have received as many as 5 answers and none of them are suitable for any of the above mentioned points. :)
==========================================================
The function looks for a true in the first column of the logical matrix, if a true is found, go to column 2 and a new row, and so on..
If the sequence is found return true if not false
set.seed(15)
m <- matrix(sample(c(F,T),30,T),ncol = 3) ; colnames(m) <- paste0("x",1:ncol(m))
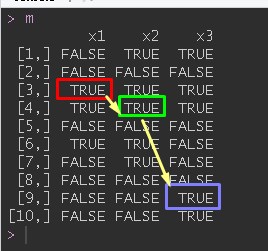
m
x1 x2 x3
[1,] FALSE TRUE TRUE
[2,] FALSE FALSE FALSE
[3,] TRUE TRUE TRUE
[4,] TRUE TRUE TRUE
[5,] FALSE FALSE FALSE
[6,] TRUE TRUE FALSE
[7,] FALSE TRUE FALSE
[8,] FALSE FALSE FALSE
[9,] FALSE FALSE TRUE
[10,] FALSE FALSE TRUE
my slow example function
find_seq <- function(m){
colum <- 1
res <- rep(FALSE,ncol(m))
for(i in 1:nrow(m)){
if(m[i,colum]==TRUE){
res[colum] <- TRUE
print(c(row=i,col=colum))
colum <- colum 1}
if(colum>ncol(m)) break
}
all(res)
}
find_seq(m)
row col
3 1
row col
4 2
row col
9 3
[1] TRUE
how to make it as fast as possible?
UPD=========================
I showed a test of only those functions that meet all three points above.
microbenchmark::microbenchmark(Jean_Claude_Arbaut_fun(m),
ThomasIsCoding_fun(m),
my_fun(m))
Unit: microseconds
expr min lq mean median uq max neval cld
Jean_Claude_Arbaut_fun(m) 2.850 3.421 4.36179 3.9915 4.5615 27.938 100 a
ThomasIsCoding_fun(m) 14.824 15.965 17.92030 16.5350 17.1050 101.489 100 b
my_fun(m) 23.946 24.517 25.59461 25.0880 25.6580 42.192 100 c
CodePudding user response:
Update
If you are pursuing the speed, you can try the following base R solution
TIC_fun <- function(m) {
p <- k <- 1
nr <- nrow(m)
nc <- ncol(m)
repeat {
if (p > nr) {
return(FALSE)
}
found <- FALSE
for (i in p:nr) {
if (m[i, k]) {
# print(c(row = i, col = k))
p <- i 1
k <- k 1
found <- TRUE
break
}
}
if (!found) {
return(FALSE)
}
if (k > nc) {
return(TRUE)
}
}
}
and you will see
Unit: microseconds
expr min lq mean median uq max neval
my_fun(m) 18.600 26.3010 41.46795 41.5510 44.3010 121.302 100
TIC_fun(m) 10.201 14.1515 409.89394 22.6505 24.4005 38906.601 100
Previous Answer
You can try the code below
lst <- with(as.data.frame(which(m, arr.ind = TRUE)), split(row, col))
# lst <- apply(m, 2, which)
setNames(
stack(
setNames(
Reduce(function(x, y) y[y > x][1],
lst,
init = -Inf,
accumulate = TRUE
)[-1],
names(lst)
)
),
c("row", "col")
)
which gives
row col
1 3 1
2 4 2
3 9 3
A more interesting implementation might be using the recursions (just for fun, but not recommanded due to the inefficiency)
f <- function(k) {
if (k == 1) {
return(data.frame(row = which(m[, k])[1], col = k))
}
s <- f(k - 1)
for (i in (tail(s, 1)$row 1):nrow(m)) {
if (m[i, k]) {
return(rbind(s, data.frame(row = i, col = k)))
}
}
}
and which gives
> f(ncol(m))
row col
1 3 1
2 4 2
3 9 3
CodePudding user response:
If I understand the problem correctly, a single loop through the rows is enough. Here is a way to do this with Rcpp. Here I only return the true/false answer, if you need the indices, it's also doable.
library(Rcpp)
cppFunction('
bool hasSequence(LogicalMatrix m) {
int nrow = m.nrow(), ncol = m.ncol();
if (nrow > 0 && ncol > 0) {
int j = 0;
for (int i = 0; i < nrow; i ) {
if (m(i, j)) {
if ( j >= ncol) {
return true;
}
}
}
}
return false;
}')
a <- matrix(c(F, F, T, T, F, T, F, F, F, F,
T, F, T, T, F, T, T, F, F, F,
T, F, T, T, F, F, F, F, T, T), ncol = 3)
a
hasSequence(a)
In order to get also the indices, the following function returns a list, with at least one element (named 'found', true or false) and if found = true, another element, named 'indices':
cppFunction('
List findSequence(LogicalMatrix m) {
int nrow = m.nrow(), ncol = m.ncol();
IntegerVector indices(ncol);
if (nrow > 0 && ncol > 0) {
int j = 0;
for (int i = 0; i < nrow; i ) {
if (m(i, j)) {
indices(j) = i 1;
if ( j >= ncol) {
return List::create(Named("found") = true,
Named("indices") = indices);
}
}
}
}
return List::create(Named("found") = false);
}')
findSequence(a)
A few links to learn about Rcpp:
- High performance functions with Rcpp, Hadley Wickham
- Rcpp for everyone, Masaki E. Tsuda
- Interfacing R with C/C , Matteo Fasiolo
- Rcpp Gallery - Articles and code examples for the Rcpp package
You have to know at least a bit of C language (preferably C , but for a basic usage, you can think of Rcpp as C with some magic syntax for R data types). The first link explains the basics of Rcpp types (vectors, matrices and lists, how to allocate, use and return them). The other links are good complements.
CodePudding user response:
If your example is representative, we assume that nrow(m) >> ncol(m). In that case, it would be more efficient to move the interation from rows to columns:
ff = function(m)
{
i1 = 1
for(j in 1:ncol(m)) {
i1 = match(TRUE, m[i1:nrow(m), j]) i1
#print(i1)
if(is.na(i1)) return(FALSE)
}
return(TRUE)
}
CodePudding user response:
A bit ugly (cause of the <<-), but it will get the job done..
tempval <- 0
lapply(split(m, col(m)), function(x) {
value <- which(x)[which(x) > tempval][1]
tempval <<- value
return(value)
})
# $`1`
# [1] 3
#
# $`2`
# [1] 4
#
# $`3`
# [1] 9
CodePudding user response:
With accumulate:
purrr::accumulate(apply(m, 2, which), .init = -Inf, ~ min(.y[.y > min(.x)]))[-1]
# or
purrr::accumulate(apply(m, 2, which), .init = -Inf, ~ .y[.y > .x][1])[-1]
# x1 x2 x3
# 3 4 9
CodePudding user response:
Here a function that focuses on case handling. It's faster than all, hope it's right :)
f <- \(m) {
stopifnot(dim(m)[2] == 3L)
e <- nrow(m)
x1 <- if (any(xx1 <- m[, 1])) {
which.max(xx1)
} else {
NA_integer_
}
x2 <- if (is.na(x1)) {
NA_integer_
}
else if (any(xx2 <- m[(x1 1):e, 2])) {
which.max(xx2) x1
} else {
NA_integer_
}
x3 <- if (is.na(x2)) {
NA_integer_
}
else if (any(xx3 <- m[(x2 1):e, 3])) {
which.max(xx3) x2
} else {
NA_integer_
}
!anyNA(c(x1, x2, x3))
}
f(m)
# [1] TRUE
m2 <- m
m2[, 3] <- FALSE
f(m2)
# [1] FALSE
Data:
set.seed(15)
m <- matrix(sample(c(FALSE, TRUE), 30, TRUE), ncol=3)