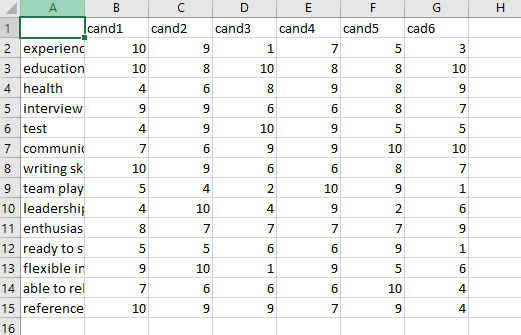
I have a dataset and the first row in it is [[10, 9, 1, 7, 5, 3].., the rows number in my dataset is 14.
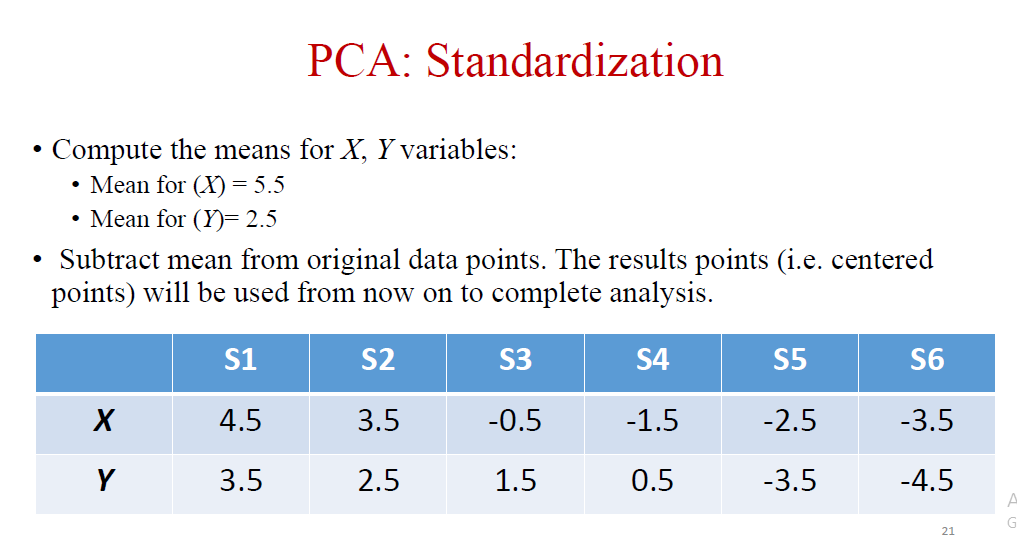
I noticed in the my lesson today, the teacher declared a mean value for each row in the dataset (Shown in the first 2 photos below) for the PCA standardization and subtracted the original data from it's own row's mean value. So I declared a c = data-data.mean() variable, and I expect the first row's mean to be (10 9 1 7 5 3) / 6 = 5.8 and c for the first index (10) to be (10 - 5.8) = 4.2, but when I printed c I found it that 10 was 3.01190476, however, when I printed out the mean I found it it wasn't for each row, but it was only 1 number which is 6.988095238095238.
My code:
dataset = pd.read_csv("cands_dataset.csv")
x = dataset.iloc[:, 1:].values
m = x.mean(dtype=np.float64);
print("x:",x) #########################
print("-------------------")
print("mean:",m) #########################
print("-------------------")
c = x - m
print("c:",c)
My output:
x: [[10 9 1 7 5 3]
[10 8 10 8 8 10]
[ 4 6 8 9 8 9]
[ 9 9 6 6 8 7]
[ 4 9 10 9 5 5]
[ 7 6 9 9 10 10]
[10 9 6 6 8 7]
[ 5 4 2 10 9 1]
[ 4 10 4 9 2 6]
[ 8 7 7 7 7 9]
[ 5 5 6 6 9 1]
[ 9 10 1 9 5 6]
[ 7 6 6 6 10 4]
[10 9 9 7 9 4]]
-------------------
mean: 6.988095238095238
-------------------
c: [[ 3.01190476 2.01190476 -5.98809524 0.01190476 -1.98809524 -3.98809524]
[ 3.01190476 1.01190476 3.01190476 1.01190476 1.01190476 3.01190476]
[-2.98809524 -0.98809524 1.01190476 2.01190476 1.01190476 2.01190476]
[ 2.01190476 2.01190476 -0.98809524 -0.98809524 1.01190476 0.01190476]
[-2.98809524 2.01190476 3.01190476 2.01190476 -1.98809524 -1.98809524]
[ 0.01190476 -0.98809524 2.01190476 2.01190476 3.01190476 3.01190476]
[ 3.01190476 2.01190476 -0.98809524 -0.98809524 1.01190476 0.01190476]
[-1.98809524 -2.98809524 -4.98809524 3.01190476 2.01190476 -5.98809524]
[-2.98809524 3.01190476 -2.98809524 2.01190476 -4.98809524 -0.98809524]
[ 1.01190476 0.01190476 0.01190476 0.01190476 0.01190476 2.01190476]
[-1.98809524 -1.98809524 -0.98809524 -0.98809524 2.01190476 -5.98809524]
[ 2.01190476 3.01190476 -5.98809524 2.01190476 -1.98809524 -0.98809524]
[ 0.01190476 -0.98809524 -0.98809524 -0.98809524 3.01190476 -2.98809524]
[ 3.01190476 2.01190476 2.01190476 0.01190476 2.01190476 -2.98809524]]
[42.28320829299149, 23.69899197734102, 18.64255211766758, 1.1547255972677037, 5.346968081158593, 8.873553933573632]
[35.09506288 19.67016334 15.47331826 0.95842225 4.43798351 7.36504976]
Teachers slide on how he did it:
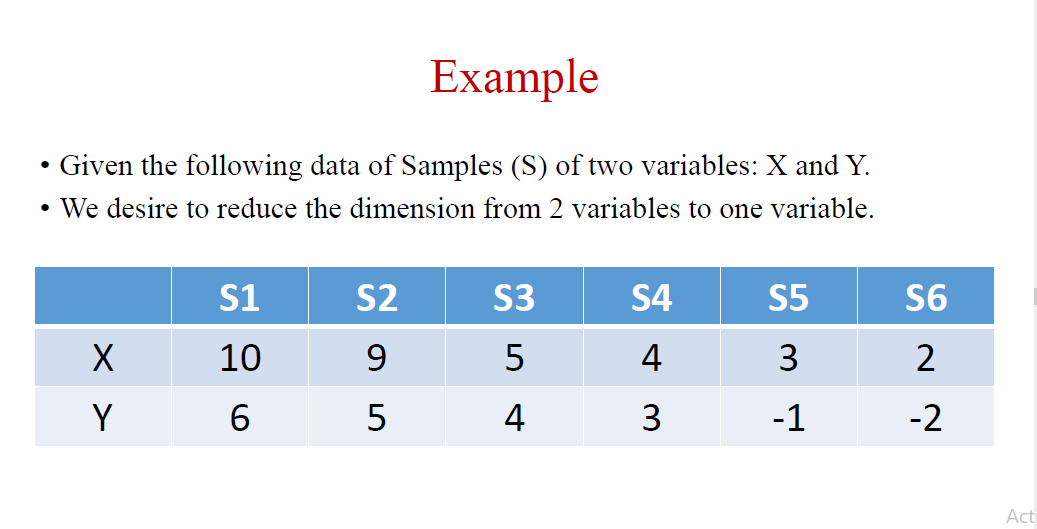 Second Slide:
Second Slide:
My dataset(Just in case if needed):
Did I understand the slides wrong? Or what's going on?
CodePudding user response:
I think when you use .iloc to subset the original dataset, you chose all the rows and all the columns after column 1, that's why the x has all the row values in it.
Please try this code if you want each mean value plus the difference row by row:
for i in range(0,len(practice)):
x = practice.iloc[i, 1: ].values
m = x.mean(dtype=np.float64);
print("x:",x) #########################
print("-------------------")
print("mean:",m) #########################
print("-------------------")
c = x - m
print("c:",c)
print('')
CodePudding user response:
Fixed it by changing the code to this m = x.mean() to m = x.mean(axis=1)