data.frame(BADAGRY_CREEK_PUBLICATION_2)
month station COPEPODA CYCLOPOIDA
1 MAY. 1 25 0
2 JUNE 1 10 5
3 JULY 1 4 0
4 AUGUST 1 5 5
5 SEPTEMBER 1 20 5
6 MAY. 2 20 0
7 JUNE 2 10 10
8 JULY 2 3 5
9 AUGUST 2 20 0
10 SEPTEMBER 2 40 0
11 MAY. 3 35 5
12 JULY 3 5 0
13 JUNE 3 25 5
14 AUGUST 3 35 5
15 SEPTEMBER 3 10 5
JUVENILE.STAGES Number.of.Species..S.
1 15 5
2 35 6
3 0 1
4 10 4
5 15 5
6 15 5
7 5 4
8 0 2
9 10 4
10 25 6
11 20 6
12 5 2
13 30 6
14 0 3
15 30 6
Number.of.Individuals..N.
1 40
2 50
3 4
4 20
5 40
6 35
7 25
8 8
9 30
10 75
11 60
12 10
13 60
14 40
15 50
was hoping tp plot stations on the x axis, with the count of each variable reflecting on the bars along the y axis. i tried this code
ggplot(BADAGRY_CREEK_PUBLICATION_2, aes(x=station)) geom_bar() [this was the bar plotted which doesnot reflect the data gathered]
the result was not well plotted
CodePudding user response:
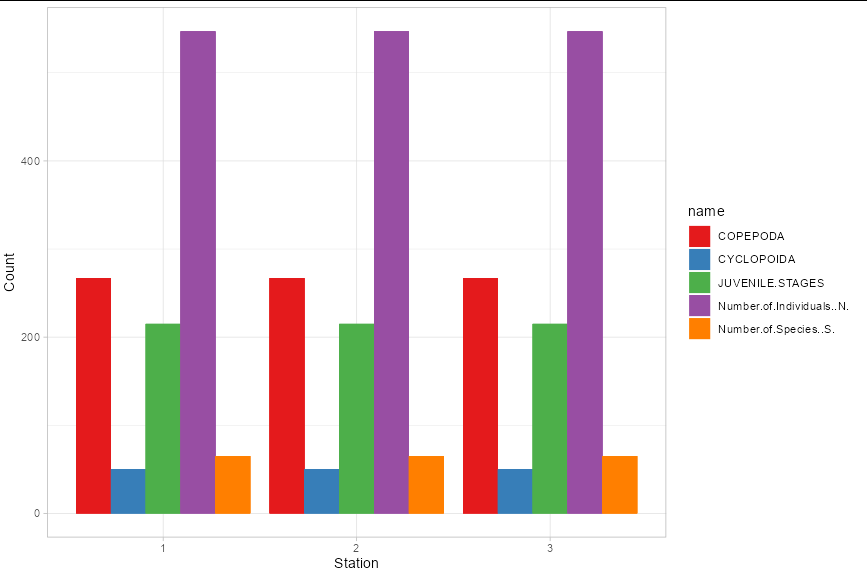
It's difficult to tell what you are looking for here, since you also have month in your data. If you want to ignore month in your plot, you could do:
library(tidyverse)
BADAGRY_CREEK_PUBLICATION_2 %>%
pivot_longer(-c(1:2)) %>%
group_by(name) %>%
summarise(value = sum(value), station = station) %>%
ggplot(aes(x = factor(station), y = value, fill = name))
geom_col(position = "dodge")
scale_fill_brewer(palette = "Set1")
theme_light()
labs(x = "Station", y = "Count")
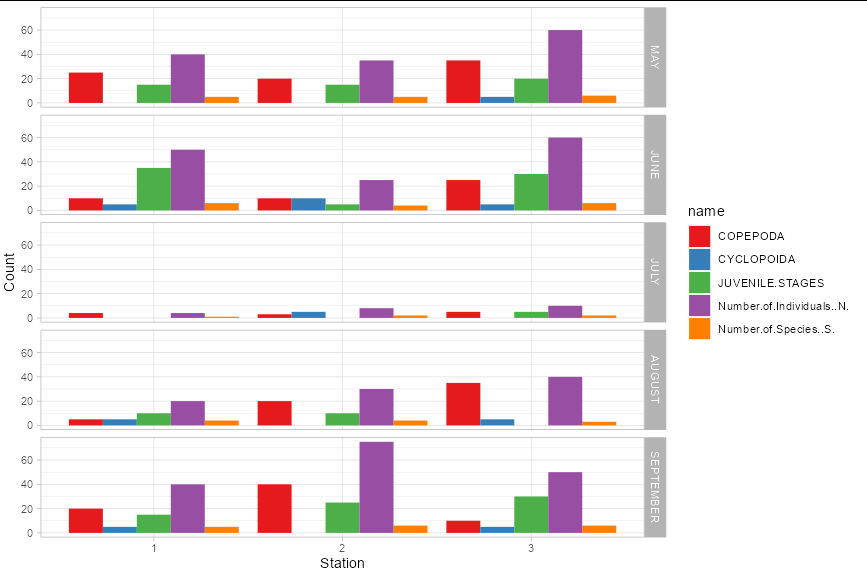
To include month, you probably need facets:
BADAGRY_CREEK_PUBLICATION_2 %>%
pivot_longer(-c(1:2)) %>%
mutate(month = factor(sub("\\.", "", month), levels = c("MAY", "JUNE",
"JULY", "AUGUST", "SEPTEMBER"))) %>%
ggplot(aes(x = factor(station), y = value, fill = name))
geom_col(position = "dodge")
facet_grid(month~.)
scale_fill_brewer(palette = "Set1")
theme_light()
labs(x = "Station", y = "Count")
Question data in reproducible format
BADAGRY_CREEK_PUBLICATION_2 <- structure(list(month = c("MAY.", "JUNE",
"JULY", "AUGUST", "SEPTEMBER",
"MAY.", "JUNE", "JULY", "AUGUST", "SEPTEMBER", "MAY.", "JULY",
"JUNE", "AUGUST", "SEPTEMBER"), station = c(1L, 1L, 1L, 1L, 1L,
2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L), COPEPODA = c(25L, 10L,
4L, 5L, 20L, 20L, 10L, 3L, 20L, 40L, 35L, 5L, 25L, 35L, 10L),
CYCLOPOIDA = c(0L, 5L, 0L, 5L, 5L, 0L, 10L, 5L, 0L, 0L, 5L,
0L, 5L, 5L, 5L), JUVENILE.STAGES = c(15L, 35L, 0L, 10L, 15L,
15L, 5L, 0L, 10L, 25L, 20L, 5L, 30L, 0L, 30L), Number.of.Species..S. = c(5L,
6L, 1L, 4L, 5L, 5L, 4L, 2L, 4L, 6L, 6L, 2L, 6L, 3L, 6L),
Number.of.Individuals..N. = c(40L, 50L, 4L, 20L, 40L, 35L,
25L, 8L, 30L, 75L, 60L, 10L, 60L, 40L, 50L)), class = "data.frame",
row.names = c("1",
"2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13",
"14", "15"))