I am following up my last question here because I got a bit greedy and I wanted to add something more to my plots so I decided I would create a new post.
So to summarize I wanted to make several plots in a loop and someone suggested using lapply which worked perfectly well for what I wanted at that time. But now, I would like to add a text reporting the corresponding value of an extra column under the title. The following code takes the suggestion of @stefan and I changed it a little bit.
So I tried my code for a subset of the dataframe for one sample and it works well but when looping with lapply I get a list of empty ggplot object...
Here is the code using a random example:
# Random example data
set.seed(123)
n <- 20
Dta <- data.frame(
sample = rep(LETTERS[seq(n)], each = 100),
x = rep(letters[seq(n)], each = 100),
value = rnorm(100 * n)
)
library(ggplot2)
library(gghighlight)
library(patchwork)
## Test with one sample
temp_df = subset(Dta, Dta$sample == "A")
letter = paste("Expected letter :",unique(temp_df$x, sep = " "))
ggplot(Dta, aes(x=value, group=sample,
color = sample))
geom_density()
labs(x = "value",
y = "Density")
theme_classic()
theme(legend.position="none",
plot.title = element_text(hjust = 0.5))
gghighlight(sample == "A",
unhighlighted_params = list(colour = "grey90"))
#Add title and text from column x
ggtitle("A")
annotate("text", x = 0.5, y = 3, label = letter, fontface = 'italic')
rm(temp_df)
# All samples with the loop
plot_fun <- function(highlight) {
temp_df = subset(Dta, Dta$sample == highlight)
letter = paste("Expected letter :",unique(temp_df$x, sep = " "))
ggplot(Dta, aes(x=value, group= sample,
color = sample))
geom_density()
labs(x = "value",
y = "Density")
theme_classic()
theme(legend.position="none",
plot.title = element_text(hjust = 0.5))
gghighlight(sample == highlight,
unhighlighted_params = list(colour = "grey90"))
#Add title and text from column x
ggtitle(highlight)
annotate("text", x = 0.5, y = 3, label = letter, fontface = 'italic')
rm(temp_df)
rm(letter)
}
Samples <- unique(Dta$sample)
# Make plots
p <- lapply(Samples, plot_fun)
This is the desired output for one sample.
And then when I remove the annotate("text", x = 0.5, y = 3, label = letter, fontface = 'italic'), it works. So adding the annotate makes the whole lapply step go wrong.
What would be your thoughts ? What am I doing wrong ? Is there maybe another way to do what I want ?
Thanks for your help people !
CodePudding user response:
The issue is that your plot function returns NULL which is the result of the rm(...) code lines at the end of your function. Remove these lines and your code works fine:
# Random example data
set.seed(123)
n <- 20
Dta <- data.frame(
sample = rep(LETTERS[seq(n)], each = 100),
x = rep(letters[seq(n)], each = 100),
value = rnorm(100 * n)
)
library(ggplot2)
library(gghighlight)
# All samples with the loop
plot_fun <- function(highlight) {
temp_df <- subset(Dta, Dta$sample == highlight)
letter <- paste("Expected letter :", unique(temp_df$x, sep = " "))
ggplot(Dta, aes(
x = value, group = sample,
color = sample
))
geom_density()
labs(
x = "value",
y = "Density"
)
theme_classic()
theme(
legend.position = "none",
plot.title = element_text(hjust = 0.5)
)
gghighlight(sample == highlight,
unhighlighted_params = list(colour = "grey90")
)
# Add title and text from column x
ggtitle(highlight)
annotate("text", x = 0.5, y = 3, label = letter, fontface = "italic")
}
Samples <- unique(Dta$sample)
# Make plots
p <- lapply(Samples, plot_fun)
p[5]
#> [[1]]
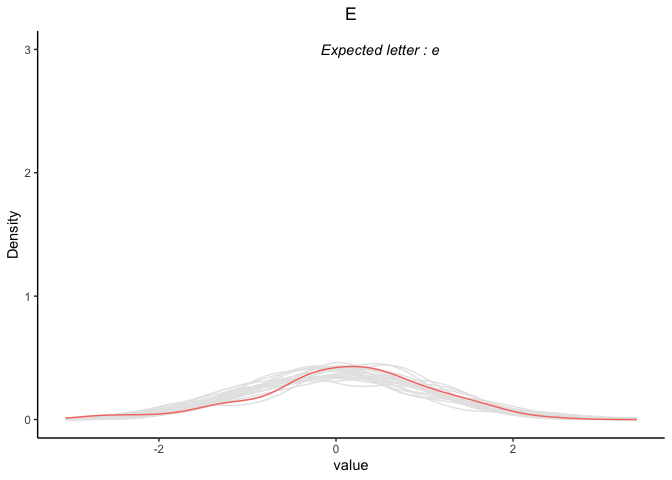
CodePudding user response:
What about trying this? Just store the plot as p, then return to the variable p at the end of the function. It would work well for the function lapply().
plot_fun <- function(highlight) {
temp_df = subset(Dta, Dta$sample == highlight)
letter = paste("Expected letter :",unique(temp_df$x, sep = " "))
p <-
ggplot(Dta, aes(x=value, group= sample,
color = sample))
geom_density()
labs(x = "value",
y = "Density")
theme_classic()
theme(legend.position="none",
plot.title = element_text(hjust = 0.5))
gghighlight(sample == highlight,
unhighlighted_params = list(colour = "grey90"))
#Add title and text from column x
ggtitle(highlight)
geom_text(aes(label = "text", x = 0.5, y = 0.3))
rm(temp_df)
rm(letter)
return(p)
}
