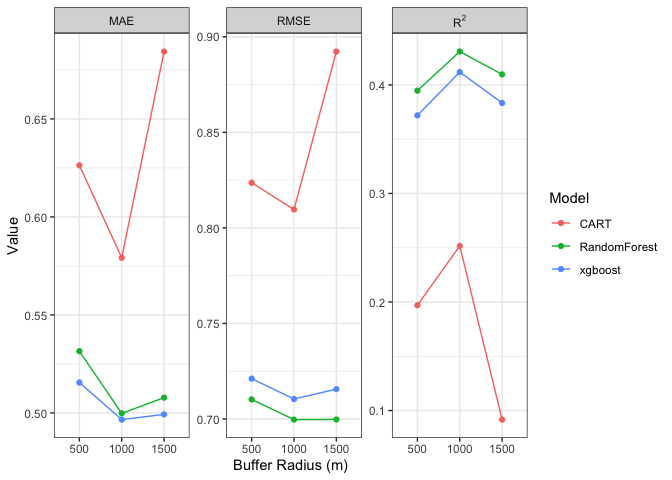
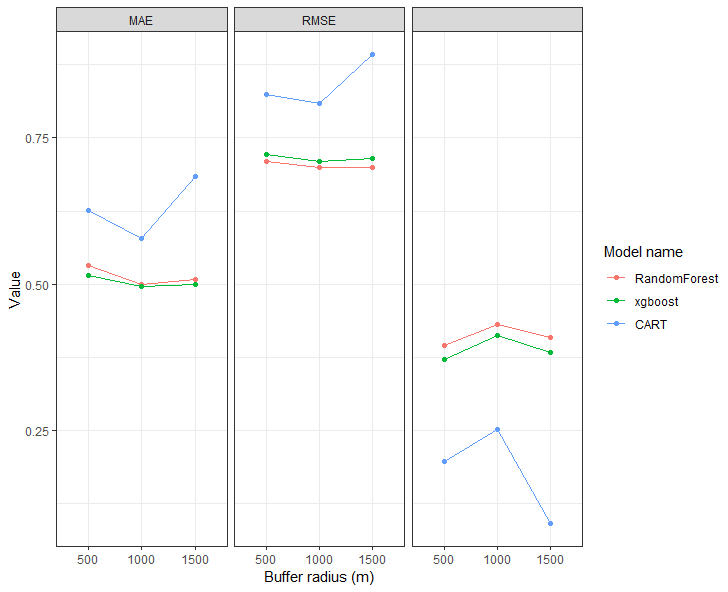
 I am using the following code to plot three sets of data and am also rearranging labels to a preferred order and making sure the 2 in R^2 is an exponent. As you can see the third panel of the facet wrap is empty. It should contain R2 (with 2 as exponent). I can't find the error that is leading to this in my code.
I am using the following code to plot three sets of data and am also rearranging labels to a preferred order and making sure the 2 in R^2 is an exponent. As you can see the third panel of the facet wrap is empty. It should contain R2 (with 2 as exponent). I can't find the error that is leading to this in my code.
library(readxl)
library(ggplot2)
library(tidyr)
error3 <- read_excel('data/ggplot_error3.xlsx')
error3 <- transform(
error3,
Metric =as.factor(Metric),
Model =as.factor(Model),
Name =as.factor(Name),
Buffer=as.factor(Buffer)
)
levels(error3$Metric)
#Set facet wrap labels
my_labeller <- as_labeller(c( MAE = 'MAE', RMSE = 'RMSE', `R^2` = 'R^2'), default = label_parsed)
# Line plot with multiple groups
ggplot(data=error3, aes(x=Buffer, y=Value, group=Model))
theme_bw()
geom_line(aes(color=Model))
geom_point(aes(color=Model))
labs(y = "Value", x = 'Buffer Radius (m)')
facet_wrap(~ Metric, nrow = 1, scales = "free_y",
labeller=my_labeller)
# Reorder line plot so R2 comes last
library(tidyverse)
library(dplyr)
error3 %>% mutate(across(Metric, factor, levels=c('MAE','RMSE','R2'))) %>%
ggplot(aes(x=Buffer, y=Value, group=Model))
geom_line(aes(color=Model))
geom_point(aes(color=Model))
theme_bw()
labs(y = "Value", x = 'Buffer radius (m)')
facet_wrap(~ Metric, nrow = 3, scales = "free_y", labeller=my_labeller)
#default level order
levels(error3$Model)
#change the order of labels
error3$Model <- factor(error3$Model, levels = c("RandomForest", "xgboost", "CART"))
#add superscript for R^2
error3 %>% mutate(across(Metric, factor, levels=c("MAE" , "RMSE", "R2"), labels = c('MAE', 'RMSE', 'R2'))) %>%
ggplot(aes(x=Buffer, y=Value, group=Model))
geom_line(aes(color=Model))
geom_point(aes(color=Model))
theme_bw()
labs(y = "Value", x = 'Buffer radius (m)')
facet_grid(~ Metric, labeller=my_labeller)
labs(color='Model name')
Code Sample:
error3 <-
structure(
list(
Buffer = structure(
c(
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L
),
.Label = c("500", "1000", "1500"),
class = "factor"
),
Area_km2 = c(
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686,
0.7854,
3.1416,
7.0686
),
Value = c(
0.626344682,
0.57916154,
0.684365417,
0.531505389,
0.49978974,
0.507811538,
0.5155205,
0.4966027,
0.4992761,
0.823620002,
0.80962234,
0.892280991,
0.710242935,
0.69967351,
0.699732972,
0.7211219,
0.7104436,
0.7156231,
0.197019417,
0.25163664,
0.09162464,
0.394737118,
0.43074958,
0.409713623,
0.3719265,
0.411827,
0.3833796
),
Metric = structure(
c(
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L
),
.Label = c("MAE", "R2", "RMSE"),
class = "factor"
),
Model = structure(
c(
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L
),
.Label = c("CART", "RandomForest", "xgboost"),
class = "factor"
),
Name = structure(
c(
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
7L,
7L,
7L,
8L,
8L,
8L,
9L,
9L,
9L,
4L,
4L,
4L,
5L,
5L,
5L,
6L,
6L,
6L
),
.Label = c(
"MAE_CART",
"MAE_rf",
"MAE_xgboost",
"R2_CART",
"R2_rf",
"R2_xgboost",
"RMSE_CART",
"RMSE_rf",
"RMSE_xgboost"
),
class = "factor"
)
),
class = "data.frame",
row.names = c(NA,-27L)
)
enter code here
CodePudding user response:
As I already mentioned in my comment the R^2 is coded as R2 in your Metric column. Hence you could fix your issue using as_labeller(c(..., R2 = "R^2")):
library(ggplot2)
library(tidyr)
library(dplyr)
my_labeller <- as_labeller(c(MAE = "MAE", RMSE = "RMSE", R2 = "R^2"), default = label_parsed)
error3 <- error3 %>% mutate(across(Metric, factor, levels = c("MAE", "RMSE", "R2")))
ggplot(data = error3, aes(x = Buffer, y = Value, group = Model))
theme_bw()
geom_line(aes(color = Model))
geom_point(aes(color = Model))
labs(y = "Value", x = "Buffer Radius (m)")
facet_wrap(~Metric,
nrow = 1, scales = "free_y",
labeller = my_labeller
)