The following code is a reproducible example based on the Swiss dataset (datasets::swiss).
My question is that I would like to plot the betas, which are the quantile regression estimates provided by the smrq() function on the y-axis, according to the tau values (the quantiles) ranging from [0:1]; but unfortunately I am not succeeding. Many thanks for the precious help, of course I can edit my post if I have forgotten anything.
Code:
library(quantreg)
library(limma)
#Generalized Functions
minimize.logcosh <- function(par, X, y, tau) {
diff <- y-(X %*% par)
check <- (tau-0.5)*diff (0.5/0.7)*logcosh(0.7*diff) 0.4
return(sum(check))
}
smrq <- function(X, y, tau){
p <- ncol(X)
op.result <- optim(
rep(0, p),
fn = minimize.logcosh,
method = 'BFGS',
X = X,
y = y,
tau = tau
)
beta <- op.result$par
return(beta)
}
run_smrq <- function(data, fml, response) {
x <- model.matrix(fml, data)[,-1]
y <- data[[response]]
X <- cbind(x, rep(1,nrow(x)))
n <- 99
betas <- sapply(1:n, function(i) smrq(X, y, tau=i/(n 1)))
return(betas)
}
#Callers
swiss <- datasets::swiss
smrq_models <- run_smrq(data=swiss, fml=Fertility~., response="Fertility")
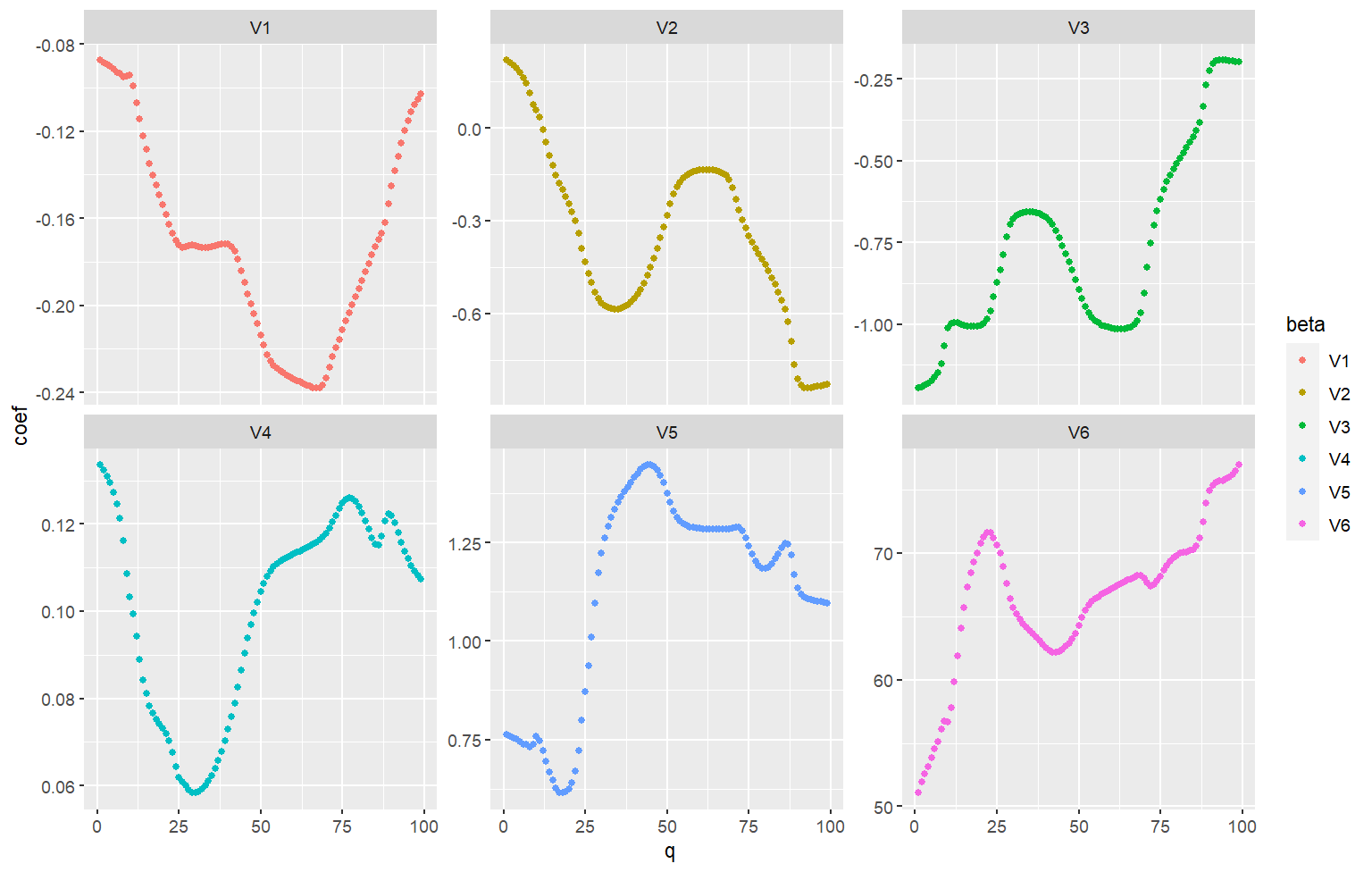
@langtang's solution gives this graphical output:
CodePudding user response:
Without making any comment on the "correctness" of the output of run_smrq(), you can try this:
library(dplyr)
library(tidyr)
library(ggplot2)
as.data.frame(t(smrq_models)) %>%
mutate(q=row_number()) %>%
pivot_longer(!q,names_to="beta",values_to = "coef") %>%
ggplot(aes(q,coef,color=beta))
geom_point()
Also, if the betas are on largely different scales, this your visualization approach may not be the most appropriate. As as starting point, you might add facet_wrap(~beta, scales="free_y")