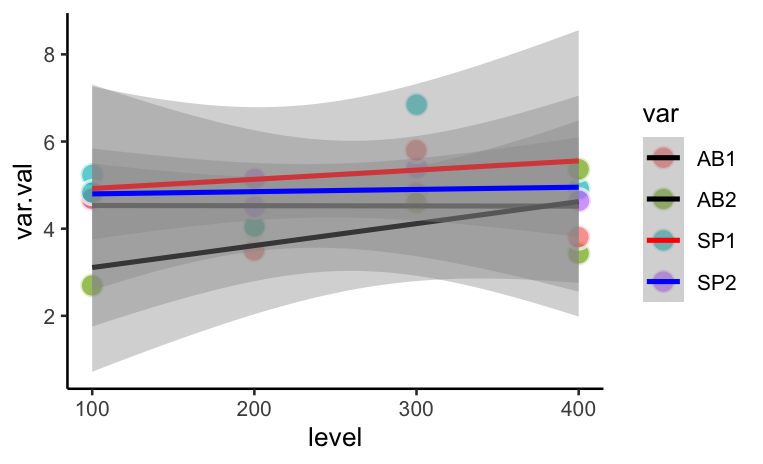
I have a dataset with species SP1 and SP2 and abiotic variables AB1 and AB2. I want to plot all of them on the graph but with modifications
Points: Display on SP1 and SP2 but NOT the AB1 and AB2 points on the graph
Lines: The AB lines should be black, dashed (see code attempt that did not work)
Confidence intervals: Only the SP lines need to have confidence intervals. The confidence intervals should have alpha = 0.2
set.seed(111) var <- rep(c("SP1","SP2","AB1","AB2"), times = 5) var.val <- rnorm(20,5,1) level <- rep(c(100,200,300,400), each = 5) df <- data.frame(var, var.val, level) ggplot(df, aes(x = level, y = var.val, col = var, group = var)) geom_point(aes(fill = var),colour="white",pch=21, size=4, stroke = 1, alpha = 0.7, position = pd) theme_classic() geom_smooth(method="lm", formula = y ~ x ) scale_linetype_manual(values = c("dashed", "dashed","solid", "solid")) scale_colour_manual(values = c("black","black","red","blue"))
CodePudding user response:
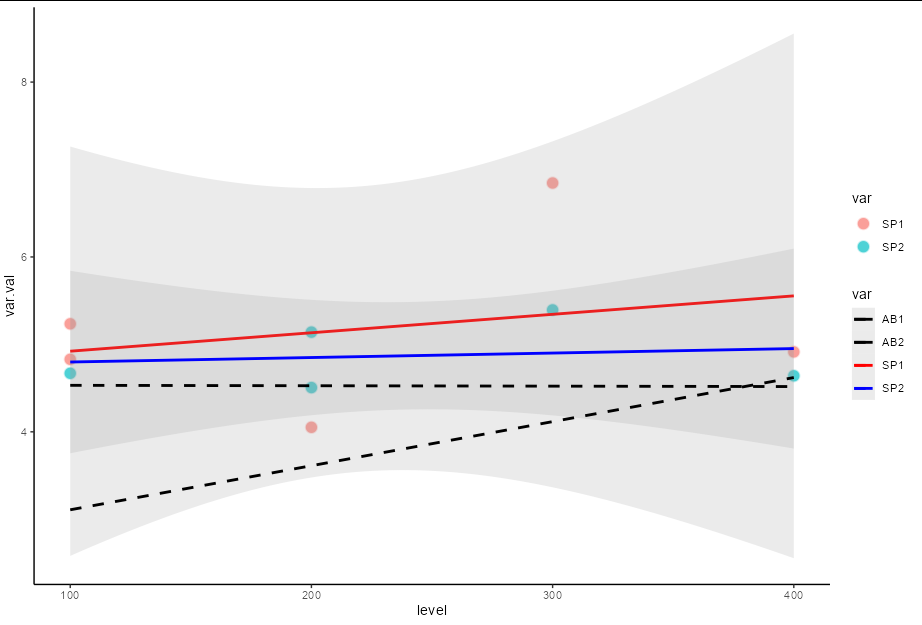
You can achieve this by passing filtered versions of the data frame to each layer:
ggplot(df, aes(x = level, y = var.val, col = var, group = var))
geom_point(aes(fill = var), colour="white",pch=21, size=4, stroke = 1,
alpha = 0.7, data = df[df$var %in% c("SP1", "SP2"),])
theme_classic()
geom_smooth(data = df[df$var %in% c("SP1", "SP2"),],
method = "lm", formula = y ~ x, alpha = 0.2)
geom_smooth(data = df[!df$var %in% c("SP1", "SP2"),],
method = "lm", formula = y ~ x, se = FALSE, linetype = 2 )
scale_linetype_manual(values = c("dashed", "dashed","solid", "solid"))
scale_colour_manual(values = c("black","black","red","blue"))
Though if AB and SP are conceptually two different variables, you might want to consider pivoting to a wider format.