I have a subset of data frame as below. I want to fill the NAs in column "age at disease" so that the age of one individual with disease be same as the sibling (identified from familyID) without disease.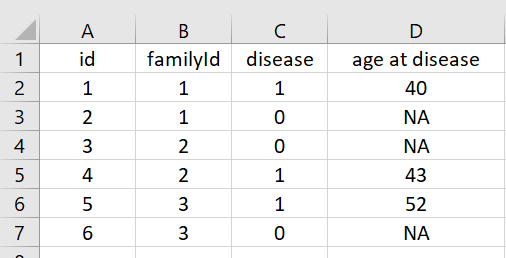
structure(list(id = c(1, 2, 3, 4, 5, 6),
familyId = c(1, 1, 2, 2, 3, 3),
disease = c(1, 0, 0, 1, 1, 0),
`age at disease` = c("40","NA", "NA", "43", "52", "NA")),
class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA, -6L))
which means that the last column "age at disease" should be: c(40,40,43,43,52,52).
CodePudding user response:
You can use the following code:
library(dplyr)
library(tidyr)
df %>%
na_if("NA") %>%
group_by(familyId) %>%
fill(`age at disease`) %>%
fill(`age at disease`, .direction = "up")
Output:
# A tibble: 6 × 4
# Groups: familyId [3]
id familyId disease `age at disease`
<dbl> <dbl> <dbl> <chr>
1 1 1 1 40
2 2 1 0 40
3 3 2 0 43
4 4 2 1 43
5 5 3 1 52
6 6 3 0 52
CodePudding user response:
If there is only a single non-NA element per group, we may also do
library(dplyr)
df1 %>%
type.convert(as.is = TRUE) %>%
group_by(familyId) %>%
mutate(`age at disease` = `age at disease`[complete.cases(`age at disease`)][1]) %>%
ungroup
-output
# A tibble: 6 × 4
id familyId disease `age at disease`
<dbl> <dbl> <dbl> <chr>
1 1 1 1 40
2 2 1 0 40
3 3 2 0 43
4 4 2 1 43
5 5 3 1 52
6 6 3 0 52
CodePudding user response:
Here is another dplyr approach:
df %>%
group_by(familyId) %>%
arrange(`age at disease`,.by_group = TRUE) %>%
mutate(`age at disease` = first(`age at disease`))
id familyId disease `age at disease`
<dbl> <dbl> <dbl> <chr>
1 1 1 1 40
2 2 1 0 40
3 4 2 1 43
4 3 2 0 43
5 5 3 1 52
6 6 3 0 52
