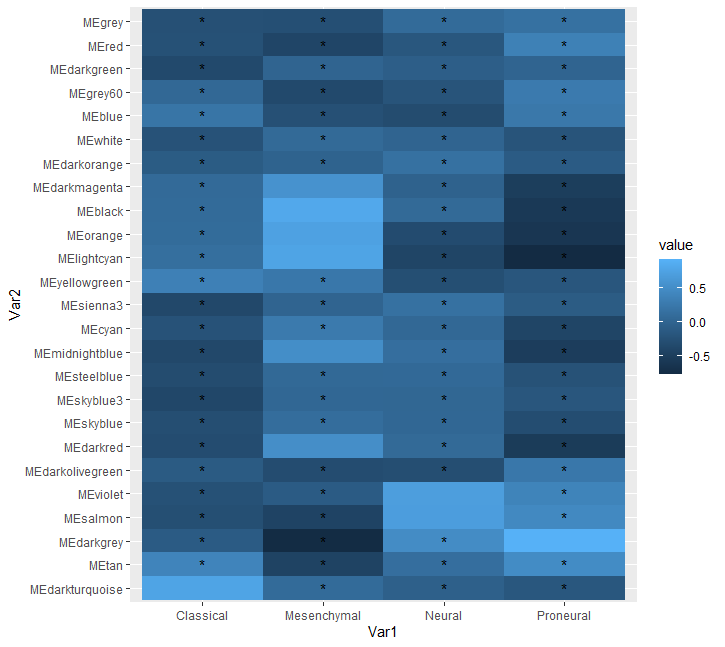
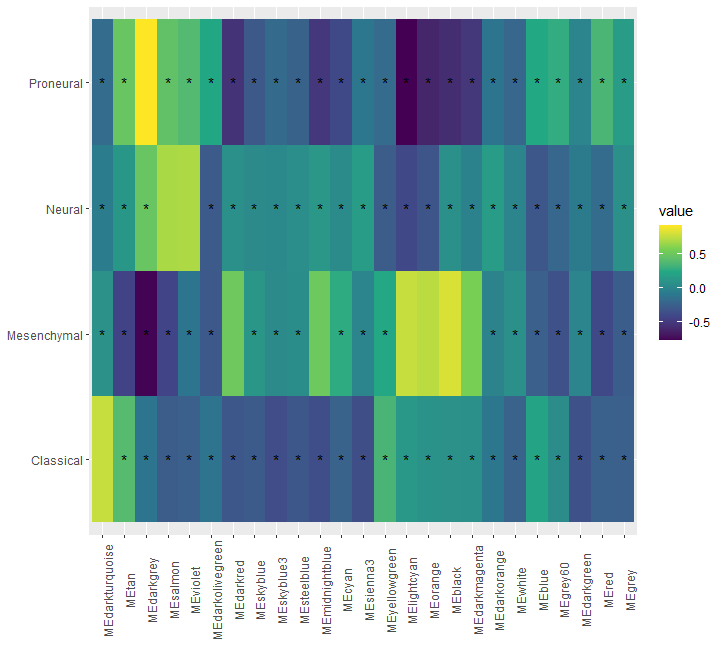
I am trying to plot a heatmap in R on a dataframe havin 4 rows and 25 columns.
dput(phenotype_MEs_spearman)
structure(c(0.775716994264046, 0.0910480716154271, -0.0514747882016944,
-0.169827441380469, 0.404562363501092, -0.435174118607051, 0.13374373005016,
0.489862481100151, -0.114284685722514, -0.761045911632707, 0.495406436748506,
0.929829361365109, -0.277485780113759, -0.429039767681859, 0.700705594355245,
0.457625938992488, -0.247512419900641, -0.113209494324045, 0.708570331437349,
0.393376052991576, -0.118035855713154, -0.305702354381165, -0.290035279717762,
0.244591643266854, -0.306940744474044, 0.518832649338805, 0.0727194182446541,
-0.53578371373029, -0.287008903928769, 0.121196630426957, 0.0386444908440733,
-0.293856049151607, -0.37281061751506, 0.0431372549019608, 0.0346989224086207,
-0.174783401732786, -0.311971104231166, 0.0589483284133727, 0.0659586723306214,
-0.237262101903185, -0.366102671178631, 0.514728682170543, 0.132315741474068,
-0.510694314445485, -0.229574963400293, 0.272292221662227, 0.0480524155806754,
-0.402311181510548, -0.367571459428325, 0.000890392876856985,
0.167470660234718, -0.106674346605227, 0.353829169366645, 0.243499652002784,
-0.27390740874073, -0.177627378980968, 0.135392516859865, 0.774555403556772,
-0.40578155374757, -0.77156502747978, 0.0965632274941801, 0.745678834569324,
-0.320267837857297, -0.614131086951304, 0.0912520699834402, 0.826493388052896,
0.0718578251373989, -0.570120239038088, 0.0827489380084959, 0.572630618955048,
-0.026514987880097, -0.506163150694794, -0.10749514003888, -0.0125206998344013,
0.16484988120095, -0.114275085799314, -0.22875176998584, 0.0727098183214534,
-0.000643194854441165, -0.206297549619603, 0.213780689754482,
-0.253296373629011, -0.312371901024792, 0.251222790217678, 0.0500611995104039,
-0.35015239878081, -0.206525547795618, 0.282588139294886, -0.346451628386973,
0.00473036215710274, -0.0702450380396957, 0.00020639834881321,
-0.242419660642715, -0.399160006719946, -0.17016103871169, 0.357323541411669,
-0.252593179254566, -0.270012239902081, 0.0845225238198095, 0.165207478340173
), .Dim = c(4L, 25L), .Dimnames = list(c("Classical", "Mesenchymal",
"Neural", "Proneural"), c("MEdarkturquoise", "MEtan", "MEdarkgrey",
"MEsalmon", "MEviolet", "MEdarkolivegreen", "MEdarkred", "MEskyblue",
"MEskyblue3", "MEsteelblue", "MEmidnightblue", "MEcyan", "MEsienna3",
"MEyellowgreen", "MElightcyan", "MEorange", "MEblack", "MEdarkmagenta",
"MEdarkorange", "MEwhite", "MEblue", "MEgrey60", "MEdarkgreen",
"MEred", "MEgrey")))
I'm using the following code:
COLORS=c("1.0","0.8","0.6","0.4","0.2","0.0","-0.2","-0.4","-0.6","-0.8","-1.0")
heatmap_spearman<-heatmap(phenotype_MEs_spearman, Colv = NA, Rowv = NA, reorderfun = function(d, w) reorder(d, w),scale="none",col= colorRampPalette(brewer.pal(8, "RdBu"))(100),ColSideColors,RowSideColors)
legend(-0.07,1, legend= COLORS[1:11],fill=colorRampPalette(brewer.pal(8, "RdBu"))(11),title="Spearman\nCorrelation")
I'm trying to put a "*" over every box in the graph where the value is <0.5, i.e., add a marker for significance. Any suggestions using other tequnues shall also be welcomed.
CodePudding user response:
I've done a heatmap before using the ggplot2 package. It's pretty easy using the
reshape2 melt() function.
Are you looking for something like this?
library(reshape2)
library(ggplot2)
b <- reshape2::melt(phenotype_MEs_spearman)
ggplot(data = b) geom_tile(aes(fill = value, x = Var1, y = Var2))
geom_text(aes(x = Var1, y = Var2, label = case_when(value < 0.5 ~ "*", TRUE ~ "") ))
You can flip the coords aswell and get rid of the x and y labs
b <- reshape2::melt(phenotype_MEs_spearman)
ggplot(data = b) geom_tile(aes(fill = value, x = Var1, y = Var2))
geom_text(aes(x = Var1, y = Var2, label = case_when(value < 0.5 ~ "*", TRUE ~ "")))
scale_fill_viridis_c()
coord_flip()
theme(axis.text.x = element_text(angle = 90))
labs(x = NULL, y = NULL)