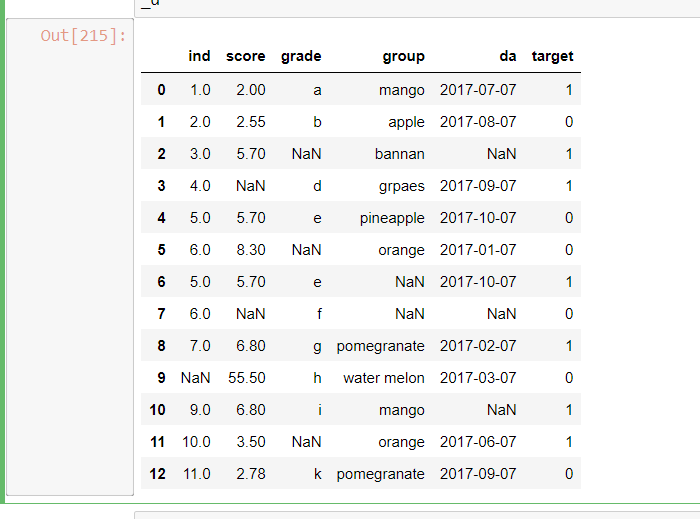
I have a small dataset, like this
_d=pd.DataFrame([
[1,2.0,'a','mango','2017-07-07',1],
[2,2.55,'b','apple','2017-08-07',0],
[3,5.7,np.nan,'bannan',np.nan,1],
[4,np.nan,'d','grpaes','2017-09-07',1],
[5,5.7,'e','pineapple','2017-10-07',0],
[6,8.3,np.nan,'orange','2017-01-07',0],
[5,5.7,'e',np.nan,'2017-10-07',1],
[6,np.nan,'f',np.nan,np.nan,0],
[7,6.8,'g','pomegranate','2017-02-07',1],
[np.nan,55.5,'h','water melon','2017-03-07',0],
[9,6.8,'i','mango',np.nan,1],
[10,3.5,np.nan,'orange','2017-06-07',1],
[11,2.78,'k','pomegranate','2017-09-07',0]
] ,columns=['ind','score','grade','group','da','target']
)
To handle NaN values and encode the category features, I used this code
y=_d['target']
x=_d.drop(['target'],axis=1)
int_columns=_d.select_dtypes(['float64','int64']).columns
obj_columns=_d.select_dtypes(['object','category']).columns
int_pipeline=Pipeline([
('impute_values',SimpleImputer(missing_values=np.nan,strategy='mean')),
('scaling',StandardScaler())
])
cat_pipeline=Pipeline([
('cat_impute',SimpleImputer(strategy='constant',fill_value='missing')),
('encoding',OneHotEncoder(drop='first'))
])
column_trans=ColumnTransformer(transformers=[
('int_p',int_pipeline,['ind', 'score']),
('cat_p',cat_pipeline,['grade', 'group'])
],remainder='passthrough')
mdl_pipeline=Pipeline([
('value_transform',column_trans)
])
transformed_data=mdl_pipeline.fit_transform(x,y)
When I run this code, I get the following error
ValueError: could not convert string to float: '2017-07-07'
The above exception was the direct cause of the following exception:
ValueError Traceback (most recent call last) Input In [253], in <cell line: 27>() 18 column_trans=ColumnTransformer(transformers=[ 19 ('int_p',int_pipeline,['ind', 'score']), 20 ('cat_p',cat_pipeline,['grade', 'group']) 21 ],remainder='passthrough') 23 mdl_pipeline=Pipeline([ 24 ('value_transform',column_trans) 25 # ,('mdl',LogisticRegression()) 26 ]) ---> 27 transformed_data=mdl_pipeline.fit_transform(x,y)
File ~\Anaconda3\lib\site-packages\sklearn\pipeline.py:434, in Pipeline.fit_transform(self, X, y, **fit_params) 432 fit_params_last_step = fit_params_steps[self.steps[-1][0]] 433 if hasattr(last_step, "fit_transform"): --> 434 return last_step.fit_transform(Xt, y, **fit_params_last_step) 435 else: 436 return last_step.fit(Xt, y, **fit_params_last_step).transform(Xt)
File ~\Anaconda3\lib\site-packages\sklearn\compose_column_transformer.py:699, in ColumnTransformer.fit_transform(self, X, y) 696 self._validate_output(Xs) 697 self._record_output_indices(Xs) --> 699 return self._hstack(list(Xs))
File ~\Anaconda3\lib\site-packages\sklearn\compose_column_transformer.py:783, in ColumnTransformer._hstack(self, Xs) 778 converted_Xs = [ 779 check_array(X, accept_sparse=True, force_all_finite=False) 780 for X in Xs 781 ] 782 except ValueError as e: --> 783 raise ValueError( 784 "For a sparse output, all columns should " 785 "be a numeric or convertible to a numeric." 786 ) from e 788 return sparse.hstack(converted_Xs).tocsr() 789 else:
ValueError: For a sparse output, all columns should be a numeric or convertible to a numeric.
The value error
ValueError: could not convert string to float: '2017-07-07'
doesnt make any sense, as I have set remainder='passthrough', In the columnTransformer, Why is my code not working
CodePudding user response:
Set sparse_threshold=0 of your ColumnTransformer. Otherwise, according to doc:
If the output of the different transformers contains sparse matrices, these will be stacked as a sparse matrix if the overall density is lower than this value. Use sparse_threshold=0 to always return dense. When the transformed output consists of all dense data, the stacked result will be dense, and this keyword will be ignored.
So it's trying to convert results of OneHotEncoder to sprase matrix but it can't since sparse matrices require numerical values (hence attempt to convert to something numerical)