I have a sample CSV file which has 2 rows.
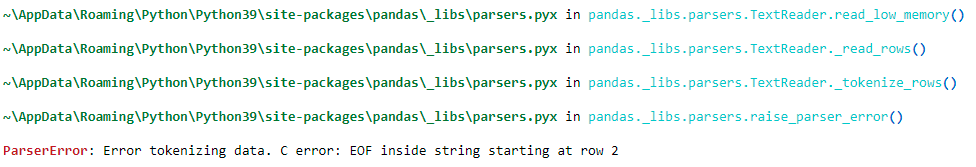
pandas.read_csv is successful if the row columns has both START & END double quotes in its columns. But if a row column has only start double quote and does not have a end double quote for the column, pandas.read_csv is failing with error, "ParserError: Error tokenizing data. C error: EOF inside string starting at row 2".
I have a requirement to filter out these bad rows to a seperate dataframe, and process the other records.
The Code I tried is :
import pandas as pd
import csv
from io import StringIO
pd.set_option('display.expand_frame_repr', False)
df = pd.read_csv('c:\\bad_data.txt', sep = '|', quotechar='"')
display(df)
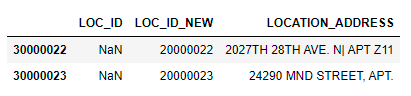
Output without bad record (the columns having both START & END quotes):
Error message when 2nd row do not have END Double quote:
I am looking for filtering this 2nd record to a seperate dataframe and process only rows with correct quotes. Please help with some suggestions/codes.
Sample data csv:
LOC_ID|LOC_ID_NEW|LOCATION_ADDRESS
30000022||20000022|"2027TH 28TH AVE. N| APT Z11"
30000023||20000023|"24290 MND STREET, APT.
CodePudding user response:
When rows are not properly formatted, it might be too difficult to use standard methods for reading them.
You could separate the bad from the good rows into two csv files first. Then you could create two data frames from these files.
original = "csvtest.csv"
ok = "csvtest_ok.csv"
bad = "csvtest_bad.csv"
def is_ok(line):
return line.endswith('\"\n')
def fix(line):
return line '\"\n'
with open(original, 'r') as o:
lines = o.readlines()
with open (ok, 'w') as o:
with open(bad, 'w') as b:
o.write(lines[0])
b.write(lines[0])
for line in lines[1:]:
if is_ok(line):
o.write(line)
else:
b.write(fix(line))
# use read_csv to create two data frames...
You might want to adapt the is_ok() and fix() methods according to your needs. They work fine for your example file.
CodePudding user response:
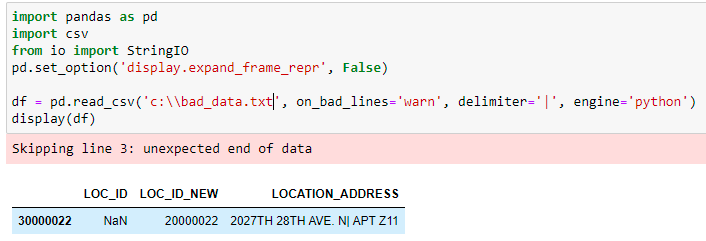
I tried with on_bad_lines='warn', while reading the CSV, and now I get a warning and also the row is skipped. Hope this helps.
df = pd.read_csv('c:\\bad_data.txt', on_bad_lines='warn', delimiter='|', engine='python')
display(df)
If you dont want the warning, you can use SKIP.
Also if you would prefer to store the warning messages to a file, then we can use redirect_stderr. Am providing a sample just in case anyone is looking for it.
import pandas as pd
from contextlib import redirect_stderr
import io
errorlist=[]
# Redirect stderr to something we can report on.
f = io.StringIO()
# with redirect_stderr(f):
with open('c:\\errors.txt', 'w') as stderr, redirect_stderr(stderr):
df = pd.read_csv('c:\\bad_data.txt', on_bad_lines='warn', delimiter='|', engine='python')
if f.getvalue():
errorlist.append("Line skipped due to parsing error : {}".format(f.getvalue()))
Thanks.