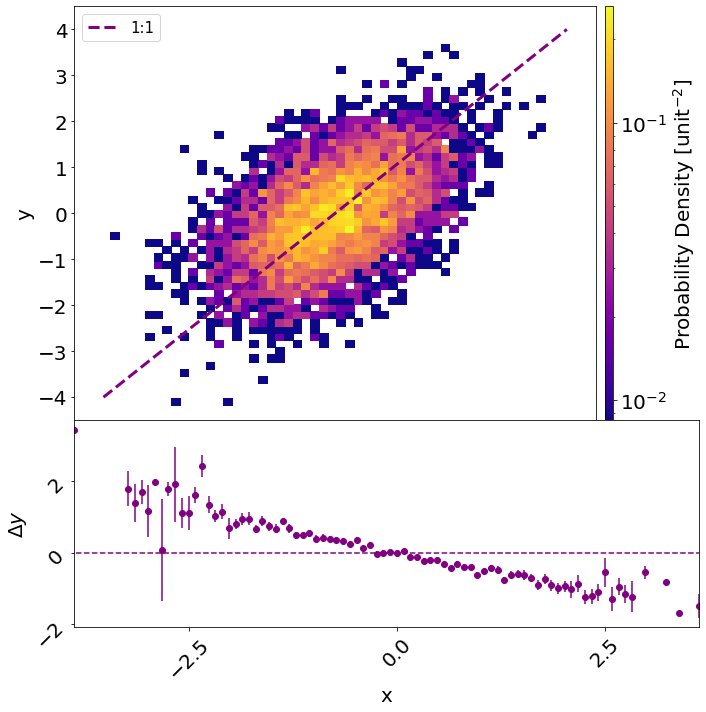
I am trying to add a GridSpec subplot to a 2D histogram in PyPlot. I want the histogram to have a colorbar. However, when I add the colorbar, the x-axes no longer match up.
How can I make the axes match?
An example of the problem I am having is replicated below. Because of the colorbar, the x-axes no longer match:
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from scipy.stats import binned_statistic, sem
import numpy as np
mean = [0, 0]
cov = [[1, 0.5], [0.5, 1]]
x, y = np.random.multivariate_normal(mean, cov, 5000).T
fig=plt.figure(figsize=(10,10))
gs = gridspec.GridSpec(2, 1, height_ratios=[2, 1])
ax0 = plt.subplot(gs[0])
h = ax.hist2d(x,
y,
density=True,
cmap="plasma",
norm=LogNorm(),
bins=100)
x_min = -4
x_max = 4
x_rng = np.linspace(x_min, x_max, 100)
h = ax0.hist2d(x,
y,
density=True,
cmap="plasma",
norm=LogNorm(),
bins=50)
ax0.plot(x_rng, x_rng, color="purple", linestyle="--", lw=3, label="1:1")
ax0.set_ylabel("y", fontsize=20)
ax0.set_ylim([x_min-0.5, x_max 0.5])
ax0.set_xlim([x_min-0.5, x_max 0.5])
cb = plt.colorbar(h[3],ax=ax0, pad = .015, aspect=50)
cb.set_label('Probability Density [unit$^{-2}$]',size=20)
cb.ax.tick_params(labelsize=20)
ax0.tick_params(axis="y", labelsize=20)
ax0.legend(fontsize=15)
ax1 = plt.subplot(gs[1], sharex = ax, aspect="auto")
y_mean, bins, _ = binned_statistic(x,
y,
statistic=np.mean,
bins = x_rng)
y_sem, bins, _ = binned_statistic(x,
y,
statistic=sem,
bins = x_rng)
x_rng_bcs = (bins[:-1] bins[1:])/2.
ax1.errorbar(x_rng_bcs, y_mean - x_rng_bcs, yerr = y_sem, color="purple", fmt="o")
ax1.errorbar(x_rng_bcs, np.zeros_like(x_rng_bcs), color="purple", linestyle="--")
ax1.tick_params(axis="x", labelsize=20, rotation=45)
ax1.tick_params(axis="y", labelsize=20, rotation=45)
ax1.set_ylabel("$\Delta y$", fontsize=20)
ax1.set_xlabel("x", fontsize=20)
fig.tight_layout()
fig.subplots_adjust(hspace=.0)
fig.show()
Which yields:
CodePudding user response:
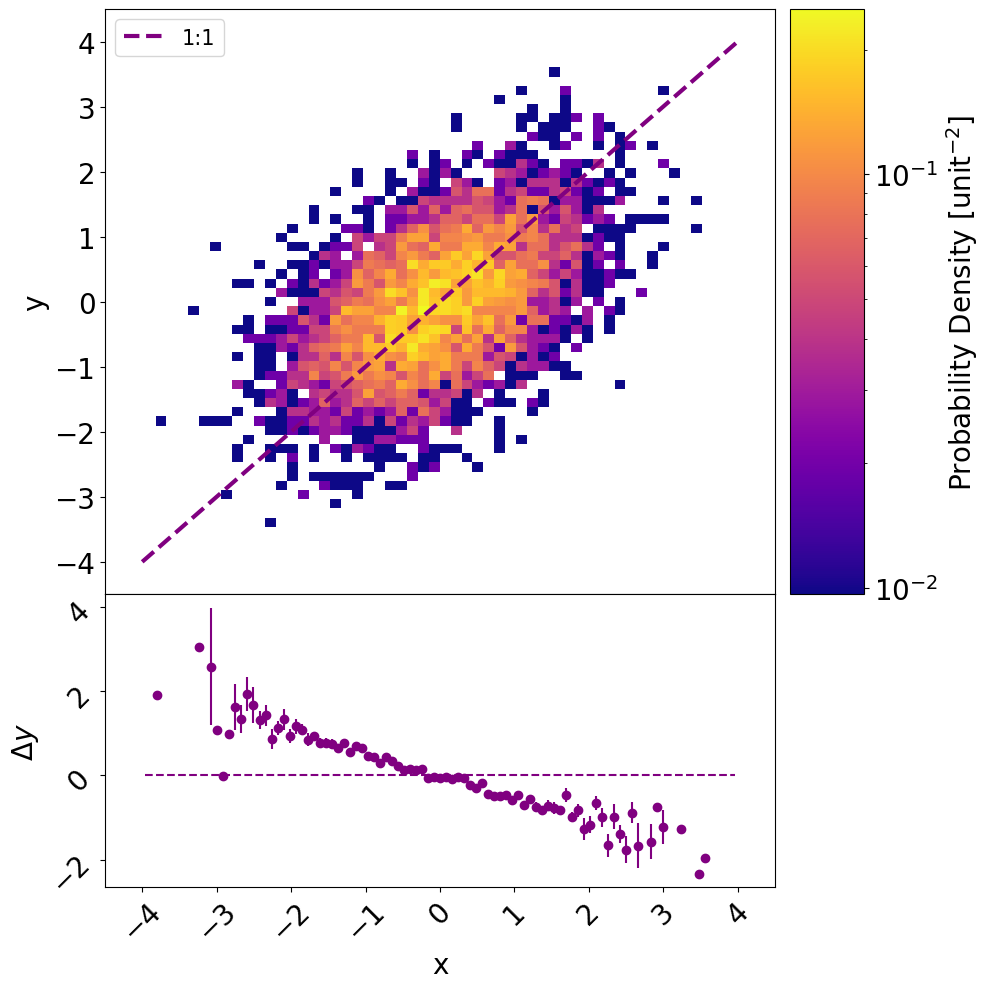
One option is to make a separate set of axes for you colorbar and use width_ratios to make it the right size.
E.g. replace
gs = gridspec.GridSpec(2, 1, height_ratios=[2, 1])
with
gs = gridspec.GridSpec(2, 2, height_ratios=[2, 1], width_ratios=[9, 1])
and then when you plot the colorbar pass the required axes as the cax keyword
ax_cb = plt.subplot(gs[0, 1])
cb = plt.colorbar(h[3], cax=ax_cb, pad = .015, aspect=50)
Doing this will give the following plot, and you can adjust the width_ratios to get the desired colorbar width.
Full code below
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from scipy.stats import binned_statistic, sem
import numpy as np
from matplotlib import gridspec
mean = [0, 0]
cov = [[1, 0.5], [0.5, 1]]
x, y = np.random.multivariate_normal(mean, cov, 5000).T
fig = plt.figure(figsize=(10, 10))
gs = gridspec.GridSpec(2, 2, height_ratios=[2, 1], width_ratios=[9, 1])
ax0 = plt.subplot(gs[0, 0])
h = ax0.hist2d(x, y, density=True, cmap="plasma", norm=LogNorm(), bins=100)
x_min = -4
x_max = 4
x_rng = np.linspace(x_min, x_max, 100)
h = ax0.hist2d(x, y, density=True, cmap="plasma", norm=LogNorm(), bins=50)
ax0.plot(x_rng, x_rng, color="purple", linestyle="--", lw=3, label="1:1")
ax0.set_ylabel("y", fontsize=20)
ax0.set_ylim([x_min - 0.5, x_max 0.5])
ax0.set_xlim([x_min - 0.5, x_max 0.5])
ax_cb = plt.subplot(gs[0, 1])
cb = plt.colorbar(h[3], cax=ax_cb, pad=0.015, aspect=50)
cb.set_label("Probability Density [unit$^{-2}$]", size=20)
cb.ax.tick_params(labelsize=20)
ax0.tick_params(axis="y", labelsize=20)
ax0.legend(fontsize=15)
ax1 = plt.subplot(gs[1, 0], sharex=ax0, aspect="auto")
y_mean, bins, _ = binned_statistic(x, y, statistic=np.mean, bins=x_rng)
y_sem, bins, _ = binned_statistic(x, y, statistic=sem, bins=x_rng)
x_rng_bcs = (bins[:-1] bins[1:]) / 2.0
ax1.errorbar(x_rng_bcs, y_mean - x_rng_bcs, yerr=y_sem, color="purple", fmt="o")
ax1.errorbar(x_rng_bcs, np.zeros_like(x_rng_bcs), color="purple", linestyle="--")
ax1.tick_params(axis="x", labelsize=20, rotation=45)
ax1.tick_params(axis="y", labelsize=20, rotation=45)
ax1.set_ylabel("$\Delta y$", fontsize=20)
ax1.set_xlabel("x", fontsize=20)
fig.tight_layout()
fig.subplots_adjust(hspace=0.0)
plt.show()
CodePudding user response:
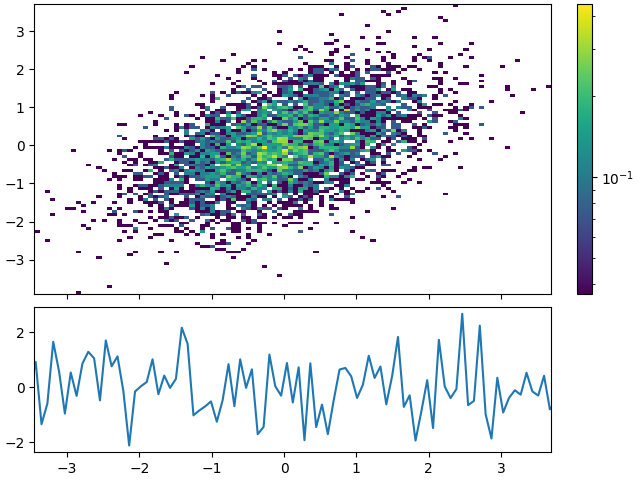
This is what constrained_layout is supposed to do:
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
import numpy as np
mean = [0, 0]
cov = [[1, 0.5], [0.5, 1]]
x, y = np.random.multivariate_normal(mean, cov, 5000).T
fig, axs =plt.subplots(2, 1, gridspec_kw={'height_ratios':[2, 1]},
sharex=True, constrained_layout=True)
ax = axs[0]
h = ax.hist2d(x, y, density=True, norm=LogNorm(), bins=100)
fig.colorbar(h[3], ax=ax)
x_rng = np.linspace(-4, 4, 100)
ax = axs[1]
ax.plot(x_rng, np.random.randn(len(x_rng)))
plt.show()