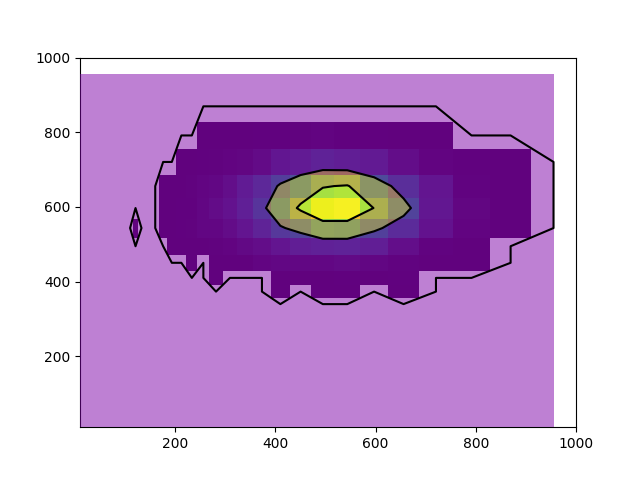
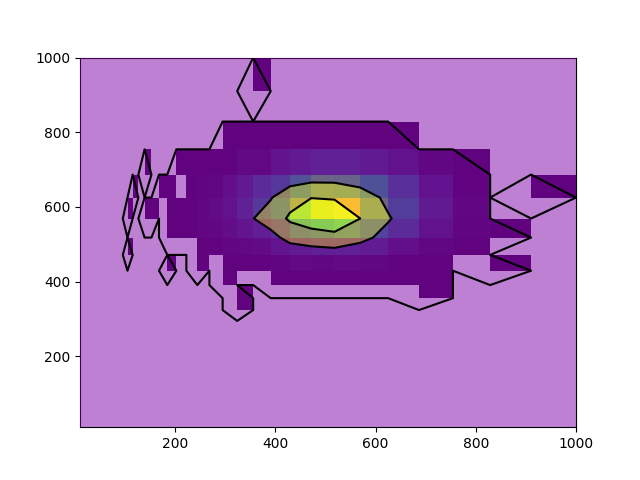
I am trying to plot contours of data that his been binned using numpy.hist2d, except the bins are set using numpy.logscale (equal binning in log space).
Unfortunately, this results in a strange behavior that I can't seem to resolve: the placement of the contours does not match the location of the points in x/y. I plot both the 2d histogram of the data, and the contours, and they do not overlap.
It looks like what is actually happening is the contours are being placed on the physical location of the plot in linear space where I expect them to be placed in log space.
It's a strange phenomenon that I think can be best described by the following plots, using identical data but binned in different ways.:
Here is a minimum working example to produce the logbinned data:
import numpy as np
import matplotlib.pyplot as plt
x = np.random.normal(loc=500, scale=100,size=10000)
y = np.random.normal(loc=600, scale=60, size=10000)
nbins = 50
bins = (np.logspace(np.log10(10),np.log10(1000),nbins),np.logspace(np.log10(10),np.log10(1000),nbins))
HH, xe, ye = np.histogram2d(x,y,bins=bins)
plt.hist2d(x,y,bins=bins,cmin=1);
grid = HH.transpose()
extent = np.array([xe.min(), xe.max(), ye.min(), ye.max()])
cs = plt.contourf(grid,2,extent=extent,extend='max',cmap='plasma',alpha=0.5,zorder=100)
plt.contour(grid,2,extent=extent,colors='k',zorder=100)
plt.yscale('log')
plt.xscale('log')
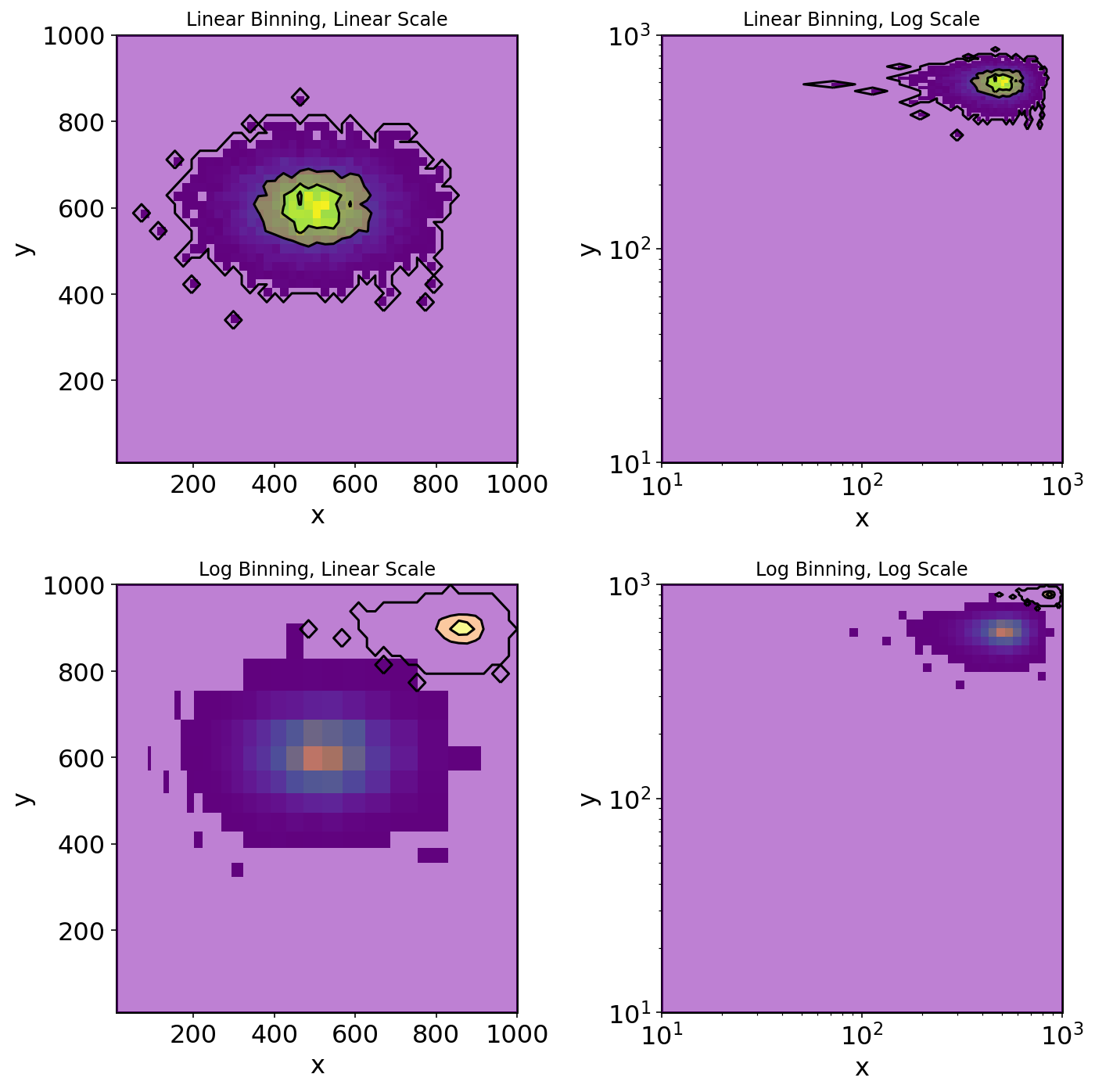
It's fairly clear what is happening -- the contour is getting misplaced do the scaling of the bins. I'd like to be able to plot the histogram and the contour here together.
If anyone has an idea of how to resolve this, that would be very helpful - thanks!
CodePudding user response:
This is your problem:
cs = plt.contourf(grid,2,extent=extent,...)
You are passing in a single 2d array specifying the values of the histograms, but you aren't passing the x and y coordinates these data correspond to. By only passing in extent there's no way for pyplot to do anything other than assume that the underlying grid is uniform, stretched out to fit extent.
So instead what you have to do is to define x and y components for each value in grid. You have to think a bit how to do this, because you have (n, n)-shaped data and (n 1,)-shaped edges to go with it. We should probably choose the center of each bin to associate a data point with. So we need to find the midpoint of each bin, and pass those arrays to contour[f].
Something like this:
import numpy as np
import matplotlib.pyplot as plt
rng = np.random.default_rng()
size = 10000
x = rng.normal(loc=500, scale=100, size=size)
y = rng.normal(loc=600, scale=60, size=size)
nbins = 50
bins = (np.geomspace(10, 1000, nbins),) * 2
HH, xe, ye = np.histogram2d(x, y, bins=bins)
fig, ax = plt.subplots()
ax.hist2d(x, y, bins=bins, cmin=1)
grid = HH.transpose()
# compute bin midpoints
midpoints = (xe[1:] xe[:-1])/2, (ye[1:] ye[:-1])/2
cs = ax.contourf(*midpoints, grid, levels=2, extend='max', cmap='plasma', alpha=0.5, zorder=100)
ax.contour(*midpoints, grid, levels=2, colors='k', zorder=100)
# these are a red herring during debugging:
#ax.set_yscale('log')
#ax.set_xscale('log')
(I've cleaned up your code a bit.)
Alternatively, if you want to avoid having those white strips at the top and edge, you can keep your bin edges, and pad your grid with zeros:
grid_padded = np.pad(grid, [(0, 1)])
cs = ax.contourf(xe, ye, grid_padded, levels=2, extend='max', cmap='plasma', alpha=0.5, zorder=100)
ax.contour(xe, ye, grid_padded, levels=2, colors='k', zorder=100)
This seems prettier, but if you think about your data this is less exact, because your data points are shifted with respect to the bin coordinates they correspond to. If you look closely you can see the contours being shifted with respect to the output of hist2d. You could fix this by generating geomspaces with one more final value which you only use for this final plotting step, and again use the midpoints of these edges (complete with a last auxiliary one).