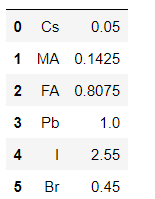
Given a molecule of the form "FA0.85MA0.15Pb(I0.85Br0.15)3". I need a dictionary with the result ("FA":"0.85", "MA":"0.15", "Pb":"1.00", "I":"2.55", "Br":"0.45").
This is obtained by multiplying the numbers (integers and floats) inside a parantheses with an integer just outside it and then separating each of the pairs. It also includes giving the value to the elements that have no compositional value mentioned (such as Pb here) should be given the value "1.00".
How can I do it with Python?
I have a bunch of such molecules and I want to create a dataframe containing all these elements and their compositions for elemental analysis.
Note: There will not be any nested brackets. All the compositions will be similar in structure to the example that I have shown above, e.g. "Cs0.05(MA0.17FA0.83)0.95Pb(I0.83Br0.17)3", "CH3NH3PbI3", "(CH3NH3)3Bi2I9".
CodePudding user response:
with regex, we can find and split the items. here are two functions to do that, the first function finds the brackets with their coefficient and the second function finds the molecules with their coefficient from the string.
so remove the brackets from the string and find and add the molecules (without brackets). then do the same work for strings that are in brackets, then multiple the molecule's coefficient in bracket coefficient, and finally add these new molecules to total molecules.
import re
def find_molecules(mol):
# find moleculars
mols = re.findall(r'([a-zA-Z] )(\d \.?\d |\d )?', mol, re.I)
# fix the non-number molecules
for i in range(len(mols)) :
mol = mols[i]
if not mol[1] :
mols[i] = (mol[0], '1')
# convert to numeric
mols = [(mol[0], float(mol[1])) for mol in mols]
return mols
def find_brackets(mol):
brks = []
# find and split brackets
for br in re.finditer(r'[\(](. )[\)](\d \.?\d |\d )', mol, re.I):
# remove bracker from string
mol = mol.replace(br[0], '')
brks.append( (br[1], float(br[2])) )
return brks, mol
def main():
molecular = "FA0.85MA0.15Pb(I0.85Br0.15)3"
data = []
# find and split the the brackets
brks, mol = find_brackets(molecular)
# add the main molecules
data.extend(find_molecules(mol))
# add the molecules that are in brackets
for br in brks:
# get molecules in bracket
mols = find_molecules(br[0])
# multiple the coefficient
mols = [(mol[0], br[1] * mol[1]) for mol in mols]
# add the mols
data.extend(mols)
print(data)
main()
CodePudding user response:
The implementation first parses the string into tokens. A small-state machine deals with missing value token and brackets&multiplier.
import re
s = "FA0.85MA0.15Pb(I0.85Br0.15)3"
# t0, t1, token type states
t0 = None; token = ''; token_list = []
for c in s ' ':
if re.match('^[a-zA-Z_] $',c): t1 = "N"
elif re.match('^[0-9.] $',c): t1 = "V"
elif re.match('^[\(] $',c): t1 = "O"
elif re.match('^[\)] $',c): t1 = "C"
else: t1 = 'E'
if not t1 == t0:
# a token is complete
if t0 == 'V': token = float(token)
token_list.append([t0,token])
if t0 == 'N' and not t1 == 'V': # insert value token of 1.0
token_list.append(['V',1.0])
if t0 == 'V' and token_list[-2][0] == 'C':
mult = token_list.pop()
l = len(token_list)
for i in range(0, l):
if token_list[l-i-1][0] == 'O': break # muliplier scope terminates
elif token_list[l-i-1][0] == 'V':
token_list[l-i-1][1] *= mult[1]
t0 = t1; token = ''
token = token c
# clean list
token_list_final = []
for item in token_list[1:]:
if item[0] == 'N' or item[0] == 'V':
token_list_final.append(item)
token_list_final
The above gives cleaned up token list:
This can be further processed into a data frame using:
V = [ item[1] for item in token_list_final[1::2]]
N = [ item[1] for item in token_list_final[ ::2]]
df = pd.DataFrame([N, V]).T
Giving: