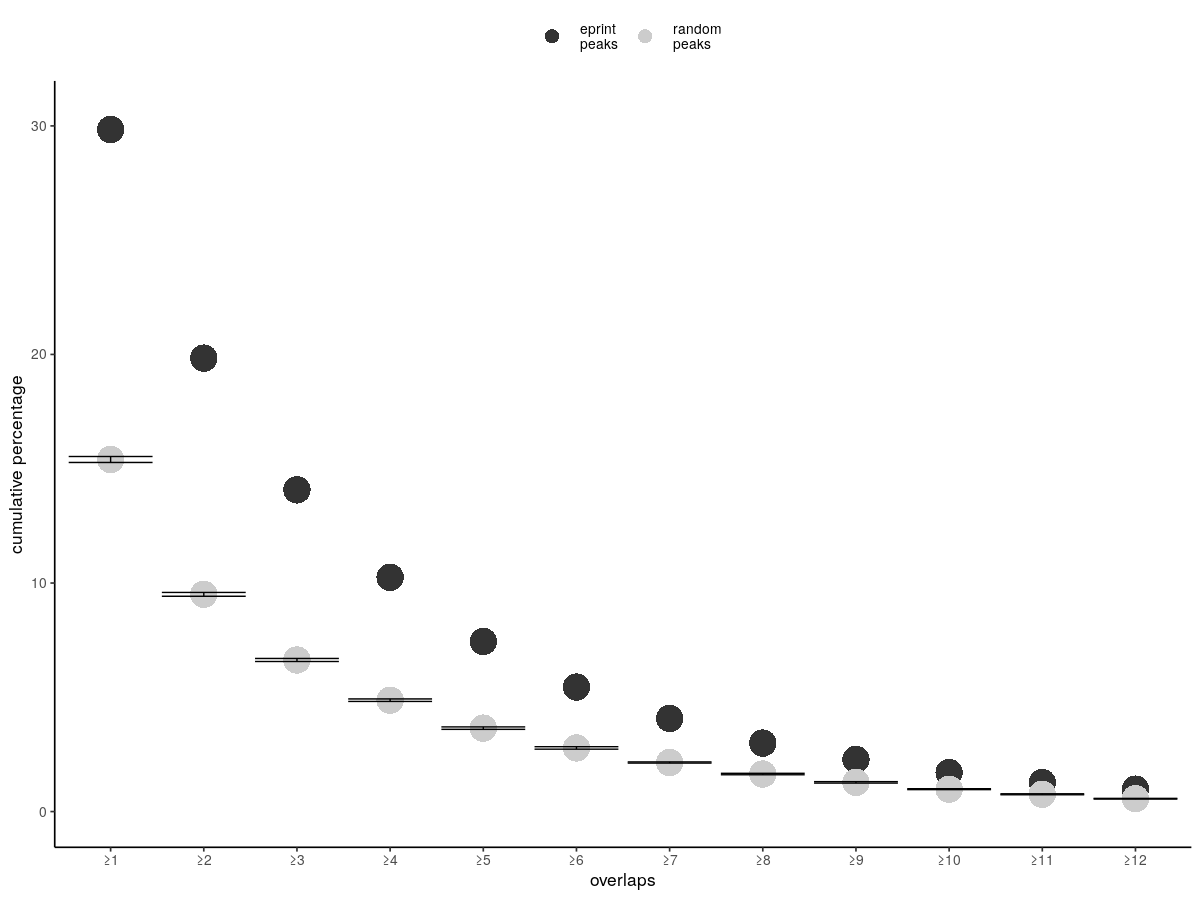
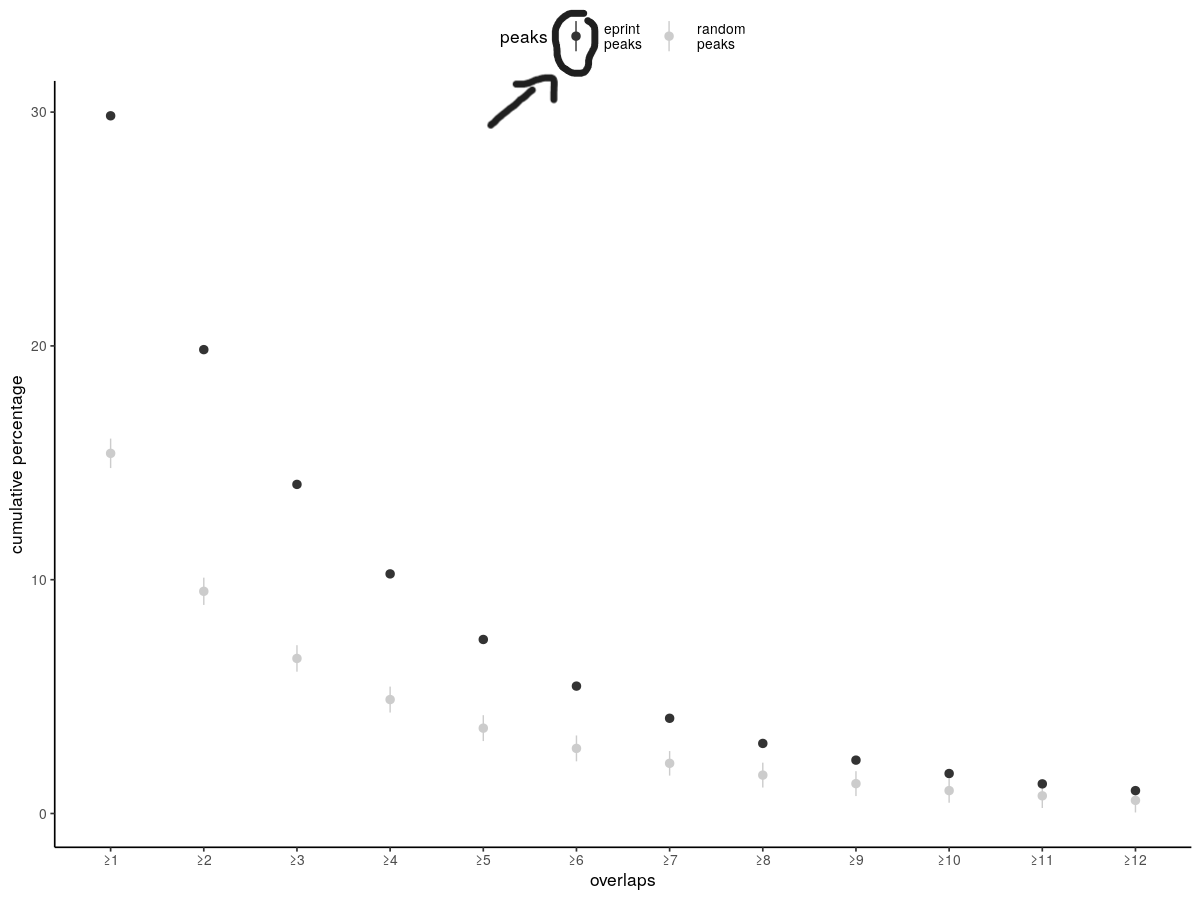
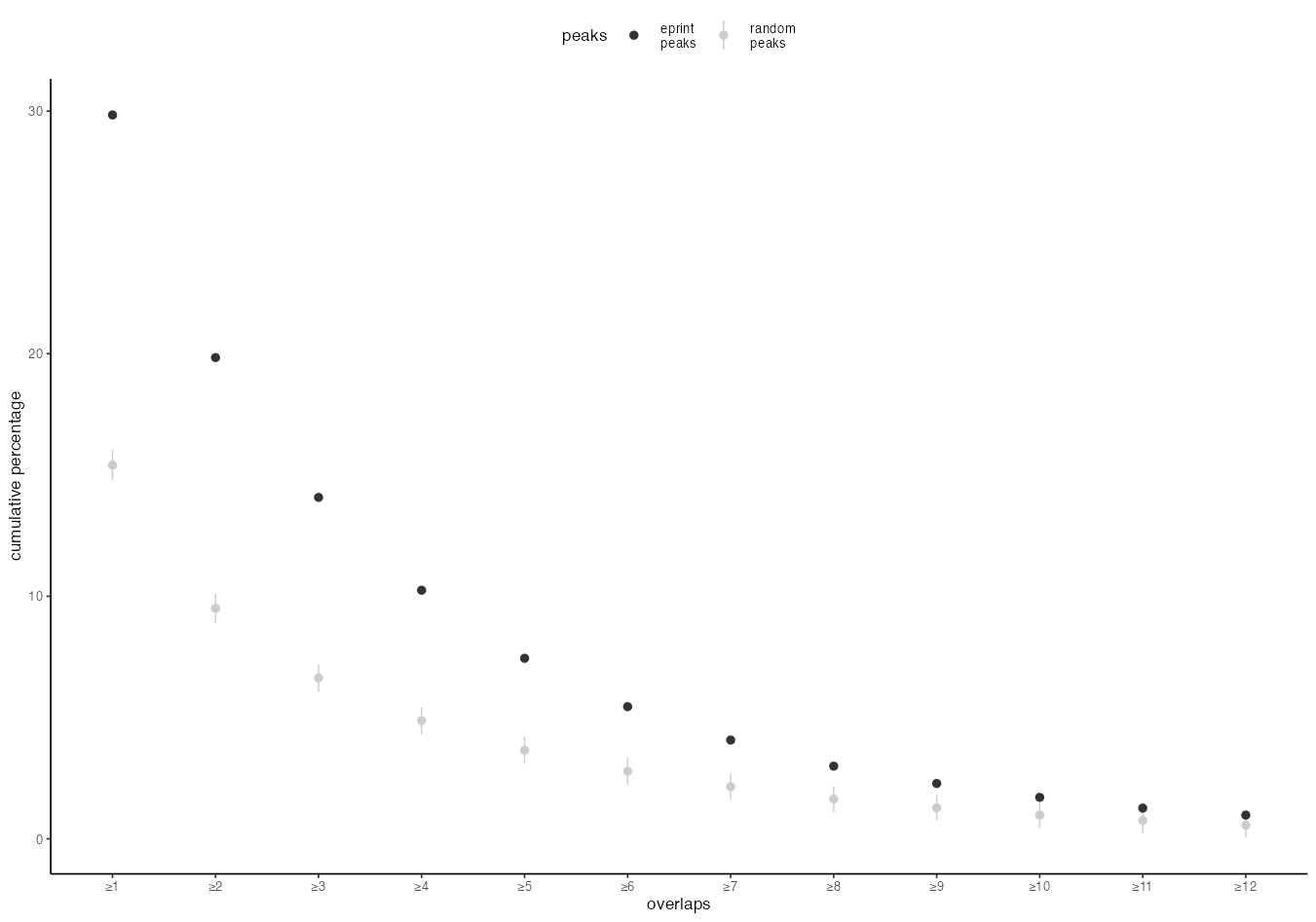
I have a dataframe as such:
dat <- data.table::data.table(
overlaps = c(1L,2L,3L,4L,5L,6L,7L,8L,9L,10L,
11L,12L,1L,2L,3L,4L,5L,6L,7L,8L,9L,10L,11L,12L),
N = c(4157L,2396L,1591L,1166L,829L,572L,
447L,297L,238L,184L,120L,90L,NA,NA,NA,NA,NA,NA,NA,NA,
NA,NA,NA,NA),
pct = c(10.0007217263695,5.76418793754661,
3.82755551278659,2.80510982269589,1.99437053431809,1.37609161113383,
1.07537229051892,0.714509105781028,0.57256958645079,
0.442658839945149,0.288690547790314,0.216517910842736,
5.90055623577914,2.87095152657789,1.75885982427862,1.22275641228556,
0.866198262032255,0.638031434348857,0.504322128003869,
0.364155269931993,0.298313566049992,0.222848843908313,
0.195119357081085,0.110664709986287),
cm_pct = c(29.8409796232588,19.8402578968894,
14.0760699593427,10.2485144465562,7.44340462386027,5.44903408954218,
4.07294247840835,2.99757018788943,2.2830610821084,
1.71049149565761,1.26783265571246,0.979142107922151,15.4040998051144,
9.50354356933526,6.63259204275737,4.87373221847875,
3.65097580619319,2.78477754416094,2.14674610981208,1.64242398180821,
1.27826871187622,0.979955145826224,0.757106301917911,
0.561986944836827),
pct_sd = c(NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,
NA,0.130441927889185,0.0834272862417102,0.0665490386787199,
0.0546826013702531,0.0526981297641897,0.0533898011751216,
0.0250810797874368,0.0320688128894919,0.0310478034379932,
0.0191658535302748,0.0211240341308091,0.0162573956365332),
peaks = as.factor(c("eprint_peaks",
"eprint_peaks","eprint_peaks","eprint_peaks",
"eprint_peaks","eprint_peaks","eprint_peaks","eprint_peaks",
"eprint_peaks","eprint_peaks","eprint_peaks",
"eprint_peaks","mean_random_peaks","mean_random_peaks",
"mean_random_peaks","mean_random_peaks",
"mean_random_peaks","mean_random_peaks","mean_random_peaks",
"mean_random_peaks","mean_random_peaks",
"mean_random_peaks","mean_random_peaks","mean_random_peaks"))
That looks like that:
overlaps N pct cm_pct pct_sd peaks
1: 1 4157 10.0007217 29.8409796 NA eprint_peaks
2: 2 2396 5.7641879 19.8402579 NA eprint_peaks
3: 3 1591 3.8275555 14.0760700 NA eprint_peaks
4: 4 1166 2.8051098 10.2485144 NA eprint_peaks
5: 5 829 1.9943705 7.4434046 NA eprint_peaks
6: 6 572 1.3760916 5.4490341 NA eprint_peaks
7: 7 447 1.0753723 4.0729425 NA eprint_peaks
8: 8 297 0.7145091 2.9975702 NA eprint_peaks
9: 9 238 0.5725696 2.2830611 NA eprint_peaks
10: 10 184 0.4426588 1.7104915 NA eprint_peaks
11: 11 120 0.2886905 1.2678327 NA eprint_peaks
12: 12 90 0.2165179 0.9791421 NA eprint_peaks
13: 1 NA 5.9005562 15.4040998 0.13044193 mean_random_peaks
14: 2 NA 2.8709515 9.5035436 0.08342729 mean_random_peaks
15: 3 NA 1.7588598 6.6325920 0.06654904 mean_random_peaks
16: 4 NA 1.2227564 4.8737322 0.05468260 mean_random_peaks
17: 5 NA 0.8661983 3.6509758 0.05269813 mean_random_peaks
18: 6 NA 0.6380314 2.7847775 0.05338980 mean_random_peaks
19: 7 NA 0.5043221 2.1467461 0.02508108 mean_random_peaks
20: 8 NA 0.3641553 1.6424240 0.03206881 mean_random_peaks
21: 9 NA 0.2983136 1.2782687 0.03104780 mean_random_peaks
22: 10 NA 0.2228488 0.9799551 0.01916585 mean_random_peaks
23: 11 NA 0.1951194 0.7571063 0.02112403 mean_random_peaks
24: 12 NA 0.1106647 0.5619869 0.01625740 mean_random_peaks
As one may notice, the pct_sd column is only available for the level mean_random_peaks of the variable peaks. I tried plot a graph using dotplot, but in order to get errorbars I found quite difficult using geom_errorbar():
Here is my first attempt:
ggplot(mapping = aes(x=factor(overlaps),y=cm_pct),data = dat)
geom_dotplot(aes(fill=peaks),colour="NA",binaxis = "y", stackdir = "centerwhole",binwidth = 1.2)
geom_errorbar(aes(ymin=cm_pct-pct_sd,ymax=cm_pct pct_sd))
scale_x_discrete(name="overlaps",breaks=seq_along(1:12),labels=paste0('\u2265',seq_along(1:12)))
theme_classic(base_size = 13)
scale_fill_grey(labels=c("eprint_peaks"="eprint\npeaks","mean_random_peaks"="random\npeaks"))
labs(y='cumulative percentage',fill=NULL)
theme(legend.position = "top",
legend.key.size = unit(1,'cm'),
)
That works great but error bars are too small because the errors in the data are small, but also the circles are too big. So overall it looks strange to me this graph.
My second attempt improves things but now the legend is awkward for eprint_peaks because there is no line at all to be drawn but nonetheless the legend prints the line.
ggplot(mapping = aes(x=factor(overlaps),y=cm_pct,colour=peaks),data = dat)
geom_pointrange(aes(ymin=cm_pct-pct_sd-.5,ymax=cm_pct pct_sd .5))
scale_x_discrete(name="overlaps",breaks=seq_along(1:12),labels=paste0('\u2265',seq_along(1:12)))
theme_classic(base_size = 13)
scale_colour_grey(labels=c("eprint_peaks"="eprint\npeaks","mean_random_peaks"="random\npeaks"))
labs(y='cumulative percentage',fill=NULL)
theme(legend.position = "top",
legend.key.size = unit(1,'cm'),
)
I tried removing the line with this command to override the shape of the legend labels but this edits both labels not only one.
guides(color = guide_legend(
override.aes=list(shape = 19)))
Is it possible to have only a dot for the legend in black and a dot line for the legend in grey ? Thanks.
CodePudding user response:
As you want to remove the line you have to override the linetypeaes:
library(ggplot2)
ggplot(mapping = aes(x=factor(overlaps),y=cm_pct,colour=peaks),data = dat)
geom_pointrange(aes(ymin=cm_pct-pct_sd-.5,ymax=cm_pct pct_sd .5))
scale_x_discrete(name="overlaps",breaks=seq_along(1:12),labels=paste0('\u2265',seq_along(1:12)))
theme_classic(base_size = 13)
scale_colour_grey(labels=c("eprint_peaks"="eprint\npeaks","mean_random_peaks"="random\npeaks"))
labs(y='cumulative percentage',fill=NULL)
theme(legend.position = "top",
legend.key.size = unit(1,'cm'),
)
guides(color = guide_legend(override.aes=list(linetype = c("blank", "solid"))))