say I have a dataframe
subject stim1 stim2 feedback
1 1003 50 51 1
2 1003 48 50 1
3 1003 49 51 1
4 1003 47 49 1
5 1003 47 46 1
6 1003 46 48 1
10 1003 50 48 1
428 1003 48 51 0
433 1003 46 50 0
434 1003 50 49 0
435 1003 54 59 0
I want to create a new column "transitive_pair" by
- group by subject (column 1),
- For every row in which feedback==0 (starting index 428, otherwise transitive_pair=NaN).
- I want to return a boolean which tells me whether there is any chain of pairings (but only those in which feedback==1) that would transitively link stim1 and stim2 values.
Working out a few examples.
row 428- stim1=48 and stim2=51
48 and 51 are not paired but 51 was paired with 50 (e.g.row 1 ) and 50 was paired with 48 (row 10) so transitive_pair[428]=True
row 433- stim 1=46 and stim2=50
46 and 48 were paired (row 6) and 48 was paired with 50 (row 2) so transitive_pair[433]=True
in row 435, stim1=54, stim2=59
there is no chain of pairs that could link them (59 is not paired with anything while feedback==1) so transitive_pair[435]=False
desired output
subject stim1 stim2 feedback transitive_pair
1 1003 50 51 1 NaN
2 1003 48 50 1 NaN
3 1003 49 51 1 NaN
4 1003 47 49 1 NaN
5 1003 47 46 1 NaN
6 1003 46 48 1 NaN
10 1003 50 48 1 NaN
428 1003 48 51 0 1
433 1003 46 50 0 1
434 1003 50 49 0 1
435 1003 54 59 0 0
any help would be greatly appreciated!!
and putting a recreateble df here
structure(list(subject = c(1003L, 1003L, 1003L, 1003L, 1003L,
1003L, 1003L, 1003L, 1003L, 1003L, 1003L), stim1 = c(50L, 48L,
49L, 47L, 47L, 46L, 50L, 48L, 46L, 50L, 54L), stim2 = c(51L,
50L, 51L, 49L, 46L, 48L, 48L, 51L, 50L, 49L, 59L), feedback = c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 0L, 0L, 0L, 0L), transitive_pair = c(NaN,
NaN, NaN, NaN, NaN, NaN, NaN, 1, 1, 1, 0)), row.names = c(1L,
2L, 3L, 4L, 5L, 6L, 10L, 428L, 433L, 434L, 435L), class = "data.frame")
CodePudding user response:
The columns "stim1" and "stim2" define an undirected graph. Create the graph for feedback == 1, get its connected components and for each row of the data.frame, check if the values of "stim1" and "stim2" belong to the same component. In the end assign NaN to the rows where feedback is 1.
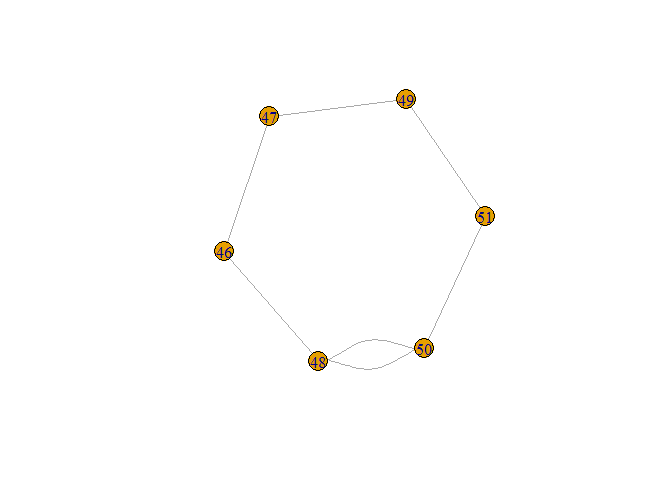
suppressPackageStartupMessages(library(igraph))
inx <- df1$feedback == 1
g <- graph_from_data_frame(df1[inx, c("stim1", "stim2")], directed = FALSE)
plot(g)
g_comp <- components(g)$membership
df1$transitive_pair_2 <- apply(df1[c("stim1", "stim2")], 1, \(x) {
i <- names(g_comp) == x[1]
j <- names(g_comp) == x[2]
if(any(i) & any(j))
g_comp[i] == g_comp[j]
else 0L
})
df1$transitive_pair_2[inx] <- NaN
df1
#> subject stim1 stim2 feedback transitive_pair transitive_pair_2
#> 1 1003 50 51 1 NaN NaN
#> 2 1003 48 50 1 NaN NaN
#> 3 1003 49 51 1 NaN NaN
#> 4 1003 47 49 1 NaN NaN
#> 5 1003 47 46 1 NaN NaN
#> 6 1003 46 48 1 NaN NaN
#> 10 1003 50 48 1 NaN NaN
#> 428 1003 48 51 0 1 1
#> 433 1003 46 50 0 1 1
#> 434 1003 50 49 0 1 1
#> 435 1003 54 59 0 0 0
Created on 2022-07-31 by the reprex package (v2.0.1)
