thanks in advance for your help
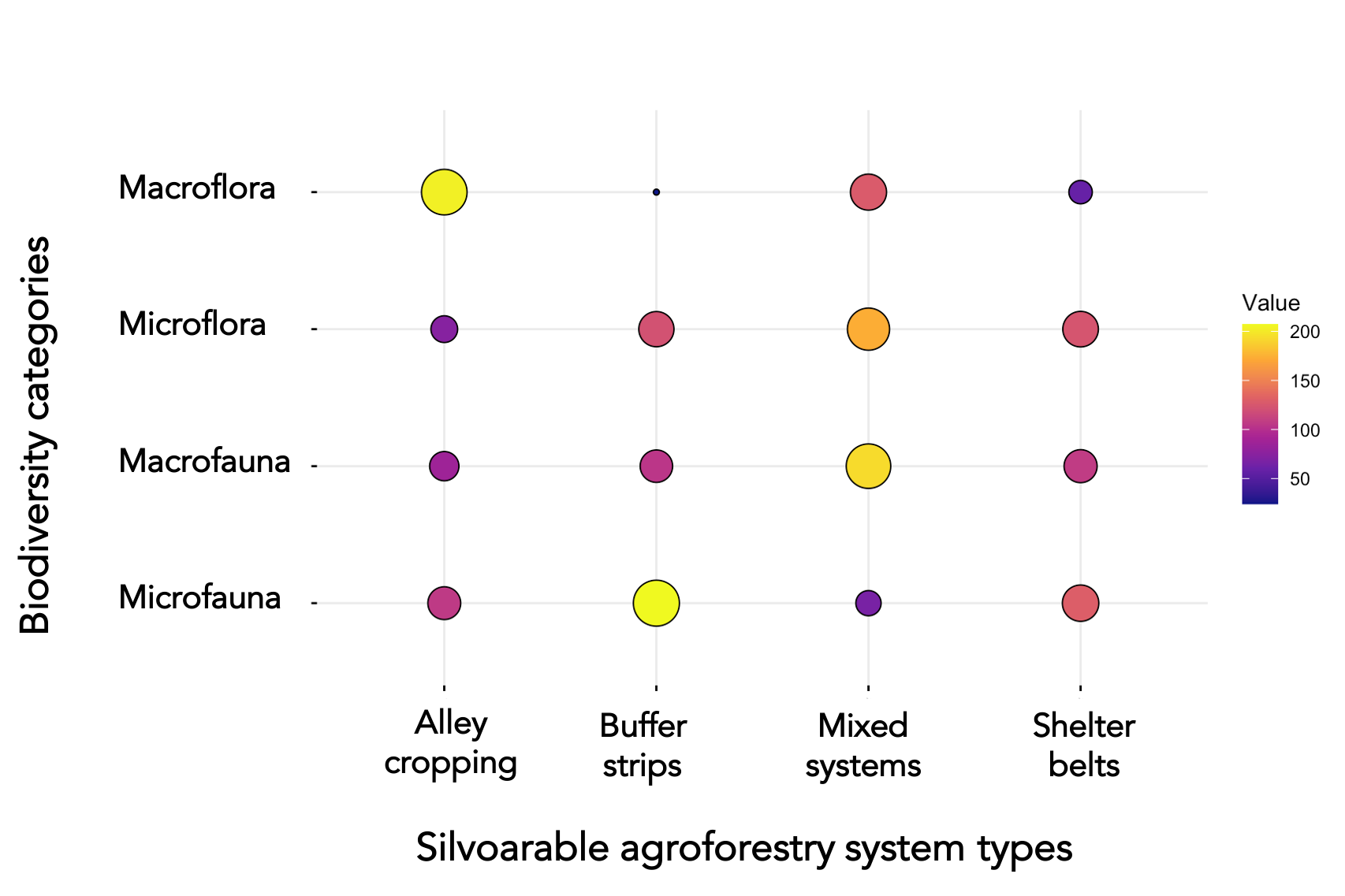
I am trying to generate a graphical matrix to illustrate the number of scientific papers reporting a certain biodiversity indicator for each type of agroforestry system.
CONTEXT
Using descriptive summary statistics to analyse a large database on silvoarable agroforestry on aspects of agroforestry systems, biodiversity and ecosystem services, as part of a systematic mapping review.
GOAL
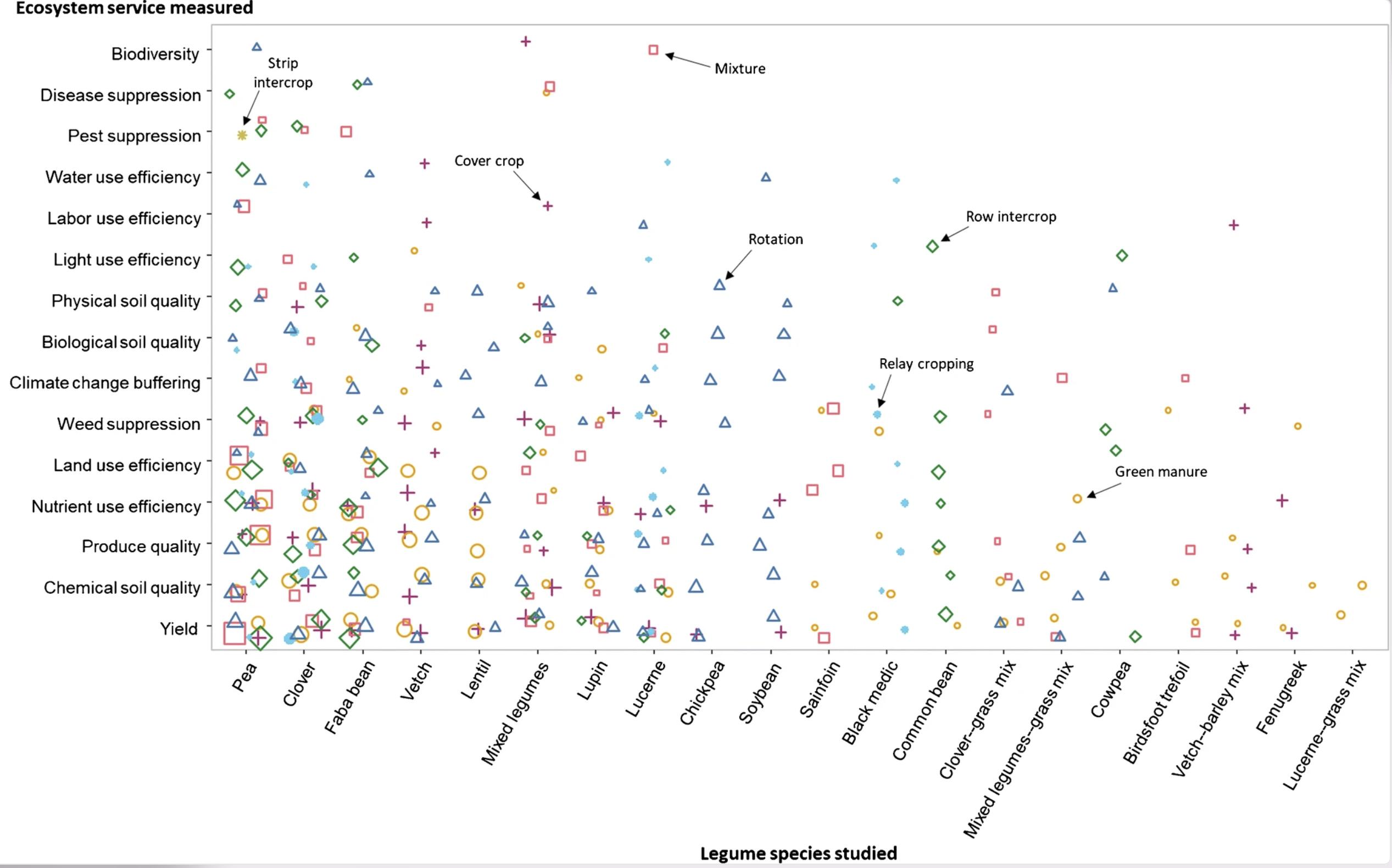
To illustrate the number of scientific papers reporting on various biodiversity indicators for each agroforestry system. To do so I would like to generate a graphical matrix (e.g. Adjacency matrix, Balloon plot), that shows the number of papers reporting on each agroforestry system and each type of biodiversity indicator. See also the attached figures.
Exemplary sketch of the desired outcome
Inspirational example from Ditzler et al. (2021)
DATA
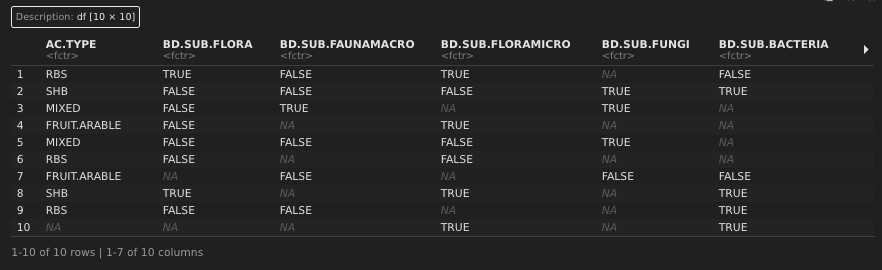
The database consists of hundreds of variables including everything from the location of the study to the specific biodiversity and ecosystem services addressed in the study. Here I have made a subset of the database that only includes the unique paper ID (ID.P), AC.TYPE (alley cropping type) and eight biodiversity variables (BD).
tibble::tribble(
~ID.P, ~ID.S, ~AC.TYPE, ~BD.SUB.FLORA, ~BD.SUB.FAUNAMACRO, ~BD.SUB.FLORAMICRO, ~BD.SUB.FUNGI, ~BD.SUB.BACTERIA, ~BD.SUB.SOILFAUNA, ~BD.SUB.SPEC, ~BD,
24, 4, "NUT.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
92, 1, "TIMBER.ARABLE", "TRUE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE", "LANDSCAPE", "TRUE",
99, 9, "SHB", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
98, 5, "SHB", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
7, 2, "TIMBER.ARABLE", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
125, 1, "BIOMASS.ARABLE", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
45, 17, "NUT.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
47, 2, "BIOMASS.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
69, 2, "TIMBER.ARABLE", "FALSE", "FALSE", "TRUE", "TRUE", "TRUE", "TRUE", "MICROBIO.BIOMASS", "TRUE",
14, 1, "TIMBER.ARABLE", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
12, 7, "BIOMASS.ARABLE", "TRUE", "FALSE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE", "TRUE",
51, 1, "BIOMASS.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
169, 1, "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
94, 7, "TIMBER.ARABLE", "TRUE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE", "LANDSCAPE", "TRUE",
49, 1, "TIMBER.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
99, 1, "SHB", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
131, 1, "MIXED", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
45, 13, "FRUIT.ARABLE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE", "FALSE",
152, 1, "BIOMASS.ARABLE", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE",
37, 29, "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "FALSE"
)
ISSUE
My issue is that I cannot convert my dataframe into the required format to generate the graphical matrix I wish to create. I have tried the xtabs() and the aggregate() function to create a contingency matrix (also called cross table), but without luck (see below). I think it is because my data is based on binary TRUE/FALSE.
CrossTab_AC.TYPE_BD <-
xtabs(~ AC.TYPE BD.SUB.FLORA BD.SUB.FAUNAMACRO BD.SUB.FAUNAMACRO BD.SUB.FLORAMICRO BD.SUB.FUNGI BD.SUB.BACTERIA BD.SUB.SOILFAUNA BD.SUB.SPEC BD,
data = BD_database) %>%
as.data.frame.table()
CrossTab_AC.TYPE_BD %>%
sample_n(20) %>%
head(10)
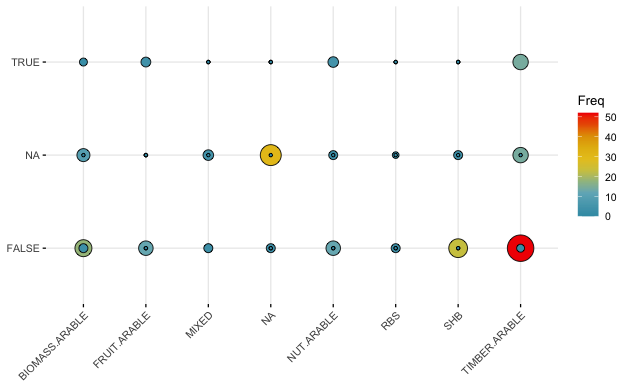
Then, I try plotting the dataframe using ggballoonplot()
ggballoonplot(CrossTab_AC.TYPE_BD, x = "AC.TYPE", y = "BD.SUB.FAUNAMACRO",
size = "Freq", fill = "Freq")
scale_fill_gradientn(colors = my_cols_pal_10)
guides(size = FALSE)
Unfortunately, this is not working..
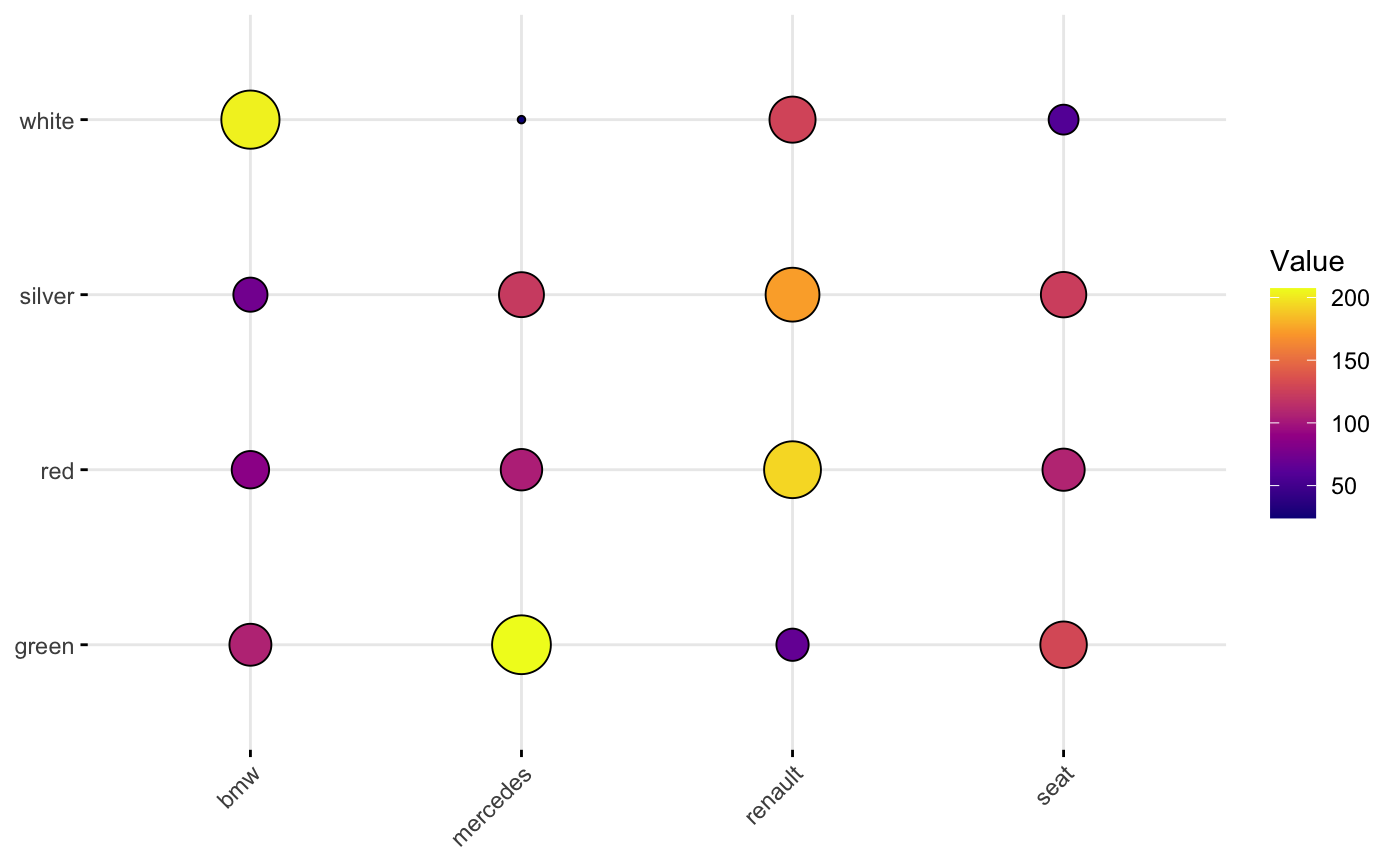
Instead, I should have a data format like what they use in there ggballoonplot example:
car_data
#> Car Color Value
#> 1 bmw red 86.2
#> 2 renault red 193.5
#> 3 mercedes red 104.2
#> 4 seat red 107.8
#> 5 bmw white 202.9
#> 6 renault white 127.7
#> 7 mercedes white 24.1
#> 8 seat white 58.8
#> 9 bmw silver 73.3
#> 10 renault silver 173.4
#> 11 mercedes silver 121.6
#> 12 seat silver 124.0
#> 13 bmw green 106.6
#> 14 renault green 66.6
#> 15 mercedes green 207.2
#> 16 seat green 129.9
ggballoonplot(car_data, x = "Car", y = "Color",
size = "Value", fill = "Value")
scale_fill_gradientn(colors = my_cols)
guides(size = FALSE)
Hence to me, it seems as if the real issue is how to convert the binary TRUE/FALSE dataset into some sort of summarised version that allows me to plot it as a graphical matrix.
I have checked on other posts here on StackOverflow, for instance, this visualizing crosstab tables with a plot in R]
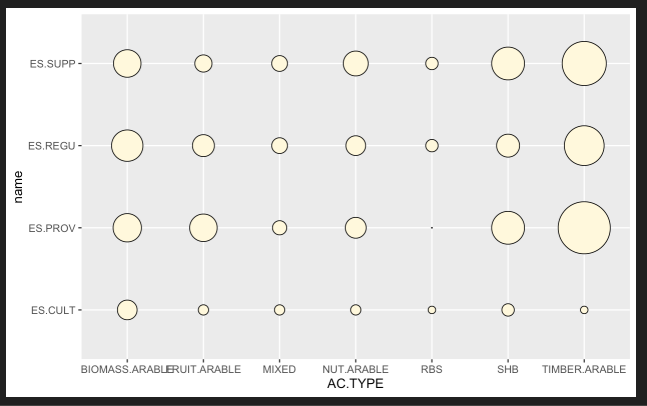
I could now simply plot it with ggplot() using these lines of code:
ggplot(try, aes(x = AC.TYPE, y = name))
geom_point(aes(size = count), shape = 21, colour = "black", fill = "cornsilk")
scale_size_area(max_size = 20, guide = "none")
Special thanks to @Quinten who has been very helpful!