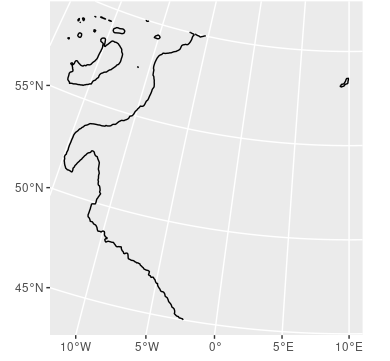
I'm trying to plot a map of the Northeast Atlantic, with specific isobaths highlighted. Given the area, I'd prefer to use the LAEA projection. However, this seems to cause geom_contour to fail.
Here is the background code:
library(tidyverse)
library(marmap)
library(rnaturalearth)
#Set boundaries
bbox <- tibble(x = c(-20, 10), y = c(45, 60))
#Add coordinates converted to LAEA
bbox <- bbox %>%
bind_cols(bbox %>%
st_as_sf(coords = c("x", "y")) %>%
st_set_crs(4326) %>% #current CRS is WSG84
st_transform(3035) %>% #transform CRS to 3035 (Lambert)
mutate(x_laea = unlist(map(geometry, 1)),
y_laea = unlist(map(geometry, 2))) %>%
st_set_geometry(NULL))
#Extract bathymetry for area of interest
nea <- fortify.bathy(getNOAA.bathy(lon1 = min(bbox$x),
lon2 = max(bbox$x),
lat1 = min(bbox$y),
lat2 = max(bbox$y),
resolution = 5))
This works nicely:
ggplot()
geom_sf(data = ne_countries(scale = "medium",
returnclass = "sf"))
geom_contour(data = nea,
aes(x = x,
y = y,
z = z),
breaks = c(-600))
coord_sf(xlim = c(min(bbox$x),
max(bbox$x)),
ylim = c(min(bbox$y),
max(bbox$y)))
But this does not (only the country layer shows up, not the nea one):
ggplot()
geom_sf(data = ne_countries(scale = "medium",
returnclass = "sf"))
geom_contour(data = nea,
aes(x = x,
y = y,
z = z))
coord_sf(crs = 3035,
xlim = c(min(bbox$x_laea),
max(bbox$x_laea)),
ylim = c(min(bbox$y_laea),
max(bbox$y_laea)))
Even if I first convert nae to LAEA first:
nea_laea <- nea %>%
st_as_sf(coords = c("x", "y")) %>%
st_set_crs(4326) %>% #current CRS is WSG84
st_transform(3035) %>% #transform CRS to 3035 (Lambert)
mutate(x = unlist(map(geometry, 1)),
y = unlist(map(geometry, 2))) %>%
st_set_geometry(NULL)
ggplot()
geom_sf(data = ne_countries(scale = "medium",
returnclass = "sf"))
geom_contour(data = nea_laea,
aes(x = x,
y = y,
z = z))
coord_sf(crs = 3035,
xlim = c(min(bbox$x_laea),
max(bbox$x_laea)),
ylim = c(min(bbox$y_laea),
max(bbox$y_laea)))
I've looked for solutions and figured that a nice way would be to extract the underlying data from the non-reprojected contour, and then plot them with the LAEA projection as geom_line:
extracted_data <- ggplot_build(ggplot()
geom_contour(data = nea,
aes(x = x, y = y, z = z),
breaks = c(-600)))$data[[1]] %>%
st_as_sf(coords = c("x", "y")) %>%
st_set_crs(4326) %>% #current CRS is WSG84
st_transform(3035) %>% #transform CRS to 3035 (Lambert)
mutate(x = unlist(map(geometry, 1)),
y = unlist(map(geometry, 2))) %>%
st_set_geometry(NULL)
This almost works, except that it seems the points are not ordered well, so the resulting plot is all scrambled:
ggplot()
geom_sf(data = ne_countries(scale = "medium",
returnclass = "sf"))
geom_line(data = extracted_data,
aes(x = x,
y = y,
group = group))
coord_sf(crs = 3035,
xlim = c(min(bbox$x_laea),
max(bbox$x_laea)),
ylim = c(min(bbox$y_laea),
max(bbox$y_laea)))
Any idea how to fix this?
Thanks a lot!
CodePudding user response:
Once you've projected your points to something other than the regular lat-long grid, you can't do a contour using geom_contour without re-interpolating onto a regular grid. help(geom_contour) explains:
Description:
ggplot2 can not draw true 3D surfaces, but you can use
‘geom_contour()’, ‘geom_contour_filled()’, and ‘geom_tile()’ to
visualise 3D surfaces in 2D. To specify a valid surface, the data
must contain ‘x’, ‘y’, and ‘z’ coordinates, and each unique
combination of ‘x’ and ‘y’ can appear at most once. Contouring
requires that the points can be rearranged so that the ‘z’ values
form a matrix, with rows corresponding to unique ‘x’ values, and
columns corresponding to unique ‘y’ values. Missing entries are
allowed, but contouring will only be done on cells of the grid
with all four ‘z’ values present. If your data is irregular, you
can interpolate to a grid before visualising using the
‘interp::interp()’ function from the ‘interp’ package (or one of
the interpolating functions from the ‘akima’ package.)
I had trouble understanding what your code was trying to do in places - you can make it more tidy by removing some of the tidyverse stuff. This:
> bbox %>% select(y_laea) %>% filter(y_laea == min(y_laea)) %>% pull()
[1] 2904046
is equivalent to this:
> min(bbox$y_laea)
[1] 2904046
and similarly for the max and X coords.
If all you want to do is get a particular contour set, use contourLines instead of trying to get values out of a plotting construct. Here's a complete example, I've tried to make this as simple as possible so it only uses sf and marmap:
Use these packages:
library(sf)
library(marmap)
set limits - we're not going to transform these so no point building a data frame with them. It also simplified extracting limits:
xr=c(-20, 10)
yr=c(45,60)
Get bathymetry as a matrix - do not "fortify" it:
bathy <- getNOAA.bathy(lon1 = min(xr),
lon2 = max(xr),
lat1 = min(yr),
lat2 = max(yr),
resolution = 5)
Now get the lat and lon coords from the dimnames. See marmap::as.xyz for where I got this from:
lon = as.numeric(rownames(bathy))
lat = as.numeric(colnames(bathy))
Now we have a matrix and coordinates so we can feed this to contourLines and get the -600 level:
c600_list = contourLines(x=lon, y=lat, z=bathy, levels=-600)
This is a list of elements with $x and $y components, so we loop over them with lapply and build st_linestring objects, then join that list of geometry primitives using st_sfc:
c600 = do.call(st_sfc,
lapply(c600_list, function(l){st_linestring(cbind(l$x, l$y))})
)
So far everything has been lat-long epsg:4326 so let's set that:
st_crs(c600) = 4326
and now we can convert to LAEA epsg:3035:
c600_laea = st_transform(c600, 3035)
You can use ggplot2 to plot:
library(ggplot2)
ggplot() geom_sf(data=c600_laea)
or use the tmap package in interactive mode to get some context and make sure we're in the right place:
library(tmap)
tmap_mode("view")
# tmap mode set to interactive viewing
tm_shape(c600_laea) tm_lines()