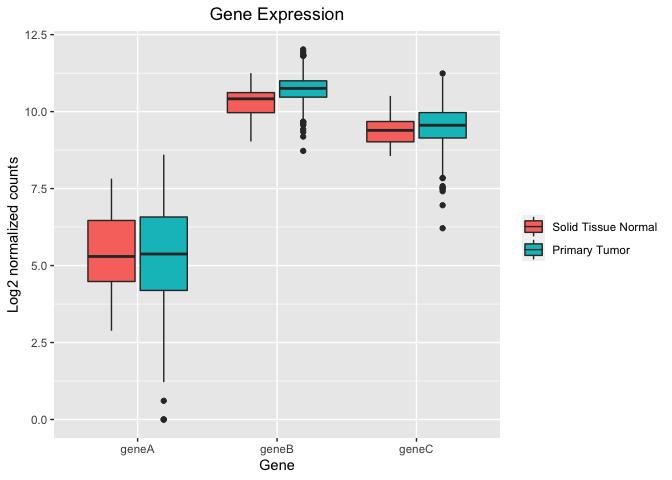
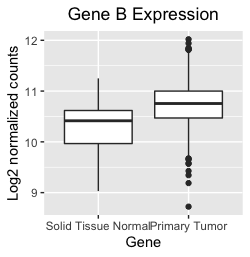
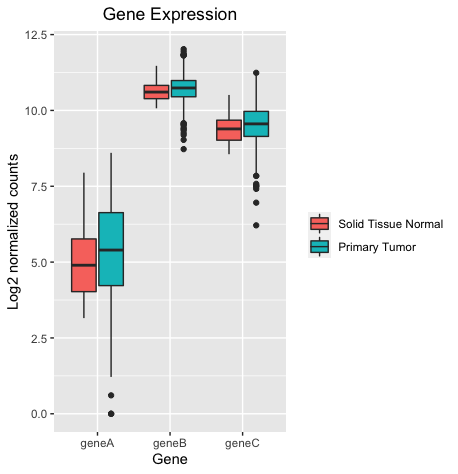
I'm using ggplot2 to make box plots to compare gene expression for genes A, B, and C in normal tissues versus tumors (about 50 normal and 500 tumor samples). The issue is that when I generate separate box plots of the individual genes, they are a bit different than if I plot all three genes together in one graph.
I'm beginning with the three separate dataframes consisting of a column of numeric expression values and a column of factors identifying tumor or normal sample type.
head(geneB)
geneB sample_type
1 12.02 Primary Tumor
2 11.94 Primary Tumor
3 11.85 Primary Tumor
4 11.84 Primary Tumor
5 11.82 Primary Tumor
6 11.82 Primary Tumor
ggplot(geneB, aes(x=sample_type, y = geneB))
geom_boxplot()
labs(title = "Gene B Expression", x= "Gene", y = "Log2 normalized counts")
theme(plot.title = element_text(hjust = 0.5))
The resulting bloxplot for gene B looks like this. Note that the entire box for the gene B tumor group is higher than the median of the normal group, and the tumor median is above the box for the normal group
Now if I combine the three dataframes together and generate a single chart with box plots for all three genes I get the following chart.
genes.df <- cbind(geneA[,1],geneB[,1],geneC)
colnames(genes.df)<-c("geneA","geneB","geneC","sample_type")
genes.df2 <- melt(genes.df, id.vars = "sample_type", variable.name = "Gene", value.name = "Normalized_Counts")
head(genes.df2)
sample_type Gene Normalized_Counts
1 Primary Tumor geneA 8.602
2 Primary Tumor geneA 8.545
3 Primary Tumor geneA 8.542
4 Primary Tumor geneA 8.420
5 Primary Tumor geneA 8.397
6 Primary Tumor geneA 8.379
>
#Combined Boxplot
ggplot(genes.df2, aes(x= Gene, y = Normalized_Counts, fill = sample_type))
geom_boxplot()
labs(title = "Gene Expression", x= "Gene", y = "Log2 normalized counts", fill = NULL)
theme(plot.title = element_text(hjust = 0.5))
Note that the gene B tumor box now extends below the normal group's median and the tumor median is not above the normal box. There are similar differences when I look closely at gene C graphed separately versus together. From manually examining the data, the separate box plots are the more correct representation of the data.
Anybody have insights or suggestions? Thanks for your help
Edits:
I suppose the issue might actually stem from an error in melting my data instead of a ggplot graphing problem. Will try to look at that more myself this afternoon.
the three dataframes are provided below
dput(geneA)
structure(list(geneA = c(8.602, 8.545, 8.542, 8.42, 8.397, 8.379,
8.286, 8.275, 8.213, 8.092, 8.081, 8.08, 8.066, 8.065, 8.061,
8.054, 8.028, 7.97, 7.966, 7.948, 7.932, 7.922, 7.921, 7.901,
7.899, 7.899, 7.881, 7.88, 7.878, 7.861, 7.855, 7.845, 7.844,
7.84, 7.828, 7.822, 7.805, 7.786, 7.779, 7.744, 7.735, 7.725,
7.715, 7.708, 7.701, 7.698, 7.698, 7.65, 7.648, 7.647, 7.64,
7.635, 7.619, 7.57, 7.562, 7.539, 7.534, 7.516, 7.48, 7.461,
7.459, 7.415, 7.401, 7.324, 7.318, 7.296, 7.288, 7.285, 7.266,
7.266, 7.262, 7.257, 7.249, 7.249, 7.232, 7.23, 7.228, 7.226,
7.212, 7.211, 7.157, 7.154, 7.142, 7.114, 7.111, 7.102, 7.102,
7.083, 7.082, 7.076, 7.075, 7.049, 7.036, 7.035, 7.034, 7.006,
6.988, 6.958, 6.945, 6.943, 6.937, 6.935, 6.926, 6.91, 6.908,
6.899, 6.886, 6.879, 6.869, 6.857, 6.852, 6.833, 6.81, 6.806,
6.801, 6.797, 6.781, 6.773, 6.768, 6.766, 6.759, 6.751, 6.744,
6.741, 6.739, 6.722, 6.721, 6.713, 6.701, 6.678, 6.671, 6.664,
6.664, 6.657, 6.656, 6.632, 6.63, 6.612, 6.606, 6.606, 6.597,
6.571, 6.547, 6.547, 6.525, 6.508, 6.492, 6.489, 6.471, 6.465,
6.464, 6.449, 6.442, 6.437, 6.411, 6.411, 6.406, 6.405, 6.4,
6.4, 6.398, 6.379, 6.37, 6.325, 6.324, 6.313, 6.308, 6.304, 6.304,
6.274, 6.271, 6.264, 6.254, 6.254, 6.238, 6.237, 6.225, 6.221,
6.21, 6.207, 6.203, 6.193, 6.193, 6.19, 6.183, 6.178, 6.151,
6.148, 6.147, 6.14, 6.132, 6.122, 6.121, 6.111, 6.107, 6.102,
6.087, 6.08, 6.08, 6.073, 6.056, 6.043, 6.028, 6.026, 6.02, 6.016,
6.014, 5.994, 5.984, 5.983, 5.965, 5.964, 5.952, 5.949, 5.934,
5.898, 5.898, 5.894, 5.881, 5.88, 5.853, 5.851, 5.84, 5.822,
5.82, 5.806, 5.802, 5.793, 5.793, 5.788, 5.782, 5.774, 5.769,
5.769, 5.759, 5.75, 5.735, 5.731, 5.72, 5.707, 5.701, 5.694,
5.694, 5.687, 5.687, 5.687, 5.668, 5.667, 5.66, 5.658, 5.647,
5.637, 5.617, 5.617, 5.614, 5.604, 5.597, 5.572, 5.57, 5.552,
5.531, 5.526, 5.498, 5.48, 5.479, 5.473, 5.469, 5.464, 5.453,
5.451, 5.449, 5.449, 5.447, 5.427, 5.42, 5.406, 5.4, 5.394, 5.385,
5.366, 5.364, 5.361, 5.353, 5.352, 5.349, 5.347, 5.338, 5.336,
5.335, 5.329, 5.318, 5.3, 5.291, 5.286, 5.283, 5.274, 5.257,
5.256, 5.255, 5.248, 5.236, 5.229, 5.226, 5.221, 5.206, 5.203,
5.194, 5.178, 5.167, 5.161, 5.147, 5.141, 5.137, 5.136, 5.12,
5.118, 5.108, 5.106, 5.103, 5.08, 5.074, 5.065, 5.063, 5.052,
5.038, 5.034, 5.029, 5.021, 5.015, 5.008, 4.993, 4.988, 4.981,
4.974, 4.97, 4.968, 4.965, 4.964, 4.956, 4.953, 4.937, 4.922,
4.921, 4.917, 4.912, 4.901, 4.897, 4.895, 4.876, 4.873, 4.851,
4.84, 4.839, 4.839, 4.829, 4.828, 4.823, 4.789, 4.788, 4.785,
4.78, 4.776, 4.753, 4.752, 4.743, 4.729, 4.699, 4.685, 4.682,
4.663, 4.65, 4.649, 4.645, 4.633, 4.63, 4.592, 4.566, 4.566,
4.566, 4.544, 4.53, 4.527, 4.524, 4.516, 4.516, 4.509, 4.504,
4.499, 4.497, 4.496, 4.495, 4.491, 4.489, 4.489, 4.47, 4.465,
4.462, 4.451, 4.439, 4.437, 4.421, 4.411, 4.4, 4.394, 4.381,
4.381, 4.379, 4.363, 4.358, 4.344, 4.338, 4.294, 4.267, 4.242,
4.239, 4.236, 4.229, 4.227, 4.22, 4.214, 4.211, 4.21, 4.208,
4.198, 4.197, 4.18, 4.177, 4.162, 4.151, 4.145, 4.142, 4.141,
4.117, 4.108, 4.1, 4.089, 4.084, 4.072, 4.069, 4.054, 4.053,
4.029, 4.028, 4.023, 4.022, 4.02, 4.017, 4.016, 4.007, 4.002,
3.993, 3.984, 3.956, 3.945, 3.929, 3.925, 3.923, 3.91, 3.899,
3.892, 3.891, 3.89, 3.889, 3.868, 3.865, 3.864, 3.863, 3.862,
3.859, 3.841, 3.841, 3.834, 3.832, 3.805, 3.783, 3.779, 3.745,
3.744, 3.735, 3.735, 3.702, 3.694, 3.684, 3.683, 3.681, 3.675,
3.652, 3.638, 3.601, 3.566, 3.557, 3.525, 3.521, 3.512, 3.511,
3.475, 3.472, 3.444, 3.441, 3.438, 3.434, 3.372, 3.342, 3.337,
3.323, 3.319, 3.318, 3.306, 3.302, 3.238, 3.238, 3.234, 3.231,
3.223, 3.211, 3.208, 3.183, 3.179, 3.154, 3.152, 3.144, 3.109,
3.103, 3.072, 3.069, 3.065, 3.063, 3.052, 3.042, 3.029, 2.987,
2.952, 2.932, 2.926, 2.917, 2.886, 2.883, 2.864, 2.808, 2.793,
2.724, 2.717, 2.65, 2.638, 2.598, 2.58, 2.559, 2.531, 2.493,
2.49, 2.463, 2.455, 2.418, 2.336, 2.332, 2.277, 2.142, 1.957,
1.58, 1.215, 0.6077, 0, 0, 0), sample_type = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 1L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L), levels = c("Solid Tissue Normal", "Primary Tumor"
), class = "factor")), row.names = c(NA, -564L), class = "data.frame")
> dput(geneB)
structure(list(geneB = c(12.02, 11.94, 11.85, 11.84, 11.82, 11.82,
11.81, 11.76, 11.64, 11.61, 11.55, 11.55, 11.53, 11.52, 11.5,
11.5, 11.5, 11.48, 11.48, 11.47, 11.46, 11.45, 11.44, 11.44,
11.42, 11.41, 11.41, 11.4, 11.4, 11.4, 11.39, 11.39, 11.39, 11.38,
11.38, 11.38, 11.37, 11.34, 11.33, 11.33, 11.32, 11.32, 11.3,
11.3, 11.3, 11.28, 11.28, 11.27, 11.27, 11.26, 11.26, 11.26,
11.26, 11.26, 11.26, 11.25, 11.25, 11.25, 11.25, 11.24, 11.24,
11.23, 11.22, 11.21, 11.21, 11.21, 11.2, 11.19, 11.19, 11.19,
11.19, 11.18, 11.18, 11.18, 11.18, 11.17, 11.16, 11.16, 11.16,
11.16, 11.16, 11.16, 11.16, 11.15, 11.15, 11.14, 11.14, 11.13,
11.12, 11.12, 11.11, 11.11, 11.11, 11.11, 11.11, 11.11, 11.11,
11.1, 11.09, 11.09, 11.09, 11.09, 11.08, 11.08, 11.07, 11.07,
11.07, 11.07, 11.06, 11.05, 11.05, 11.05, 11.05, 11.04, 11.04,
11.04, 11.04, 11.04, 11.04, 11.03, 11.03, 11.03, 11.02, 11.02,
11.01, 11.01, 11.01, 11.01, 11, 11, 11, 11, 11, 11, 11, 11, 10.98,
10.98, 10.98, 10.97, 10.97, 10.97, 10.97, 10.97, 10.97, 10.97,
10.96, 10.96, 10.96, 10.96, 10.96, 10.96, 10.96, 10.95, 10.95,
10.95, 10.95, 10.95, 10.95, 10.95, 10.94, 10.94, 10.94, 10.94,
10.93, 10.93, 10.93, 10.93, 10.93, 10.92, 10.92, 10.92, 10.92,
10.92, 10.92, 10.92, 10.91, 10.91, 10.91, 10.91, 10.91, 10.91,
10.91, 10.91, 10.9, 10.9, 10.9, 10.9, 10.9, 10.9, 10.9, 10.89,
10.89, 10.89, 10.89, 10.89, 10.89, 10.88, 10.88, 10.88, 10.88,
10.87, 10.87, 10.87, 10.87, 10.87, 10.87, 10.87, 10.86, 10.86,
10.86, 10.86, 10.86, 10.86, 10.85, 10.85, 10.85, 10.85, 10.85,
10.84, 10.84, 10.84, 10.84, 10.84, 10.84, 10.83, 10.83, 10.83,
10.83, 10.83, 10.83, 10.83, 10.82, 10.82, 10.82, 10.82, 10.82,
10.81, 10.81, 10.81, 10.81, 10.81, 10.81, 10.8, 10.8, 10.79,
10.79, 10.79, 10.79, 10.79, 10.79, 10.78, 10.78, 10.78, 10.78,
10.77, 10.77, 10.77, 10.77, 10.77, 10.77, 10.77, 10.77, 10.76,
10.76, 10.76, 10.75, 10.75, 10.75, 10.75, 10.75, 10.75, 10.74,
10.74, 10.74, 10.74, 10.74, 10.74, 10.74, 10.74, 10.73, 10.73,
10.71, 10.71, 10.71, 10.71, 10.71, 10.71, 10.71, 10.71, 10.7,
10.69, 10.69, 10.69, 10.69, 10.68, 10.68, 10.68, 10.68, 10.68,
10.68, 10.68, 10.68, 10.68, 10.67, 10.67, 10.67, 10.67, 10.67,
10.67, 10.67, 10.67, 10.67, 10.66, 10.66, 10.66, 10.66, 10.66,
10.66, 10.66, 10.66, 10.66, 10.66, 10.65, 10.65, 10.65, 10.65,
10.65, 10.64, 10.64, 10.64, 10.64, 10.64, 10.63, 10.63, 10.63,
10.63, 10.63, 10.63, 10.63, 10.62, 10.62, 10.61, 10.61, 10.61,
10.61, 10.61, 10.61, 10.61, 10.6, 10.6, 10.6, 10.6, 10.6, 10.6,
10.6, 10.6, 10.6, 10.6, 10.59, 10.59, 10.59, 10.59, 10.58, 10.58,
10.58, 10.58, 10.58, 10.57, 10.57, 10.57, 10.57, 10.57, 10.57,
10.56, 10.56, 10.56, 10.56, 10.55, 10.55, 10.55, 10.55, 10.54,
10.53, 10.53, 10.53, 10.52, 10.52, 10.52, 10.52, 10.51, 10.51,
10.51, 10.5, 10.5, 10.49, 10.49, 10.49, 10.49, 10.48, 10.48,
10.48, 10.48, 10.48, 10.47, 10.47, 10.47, 10.47, 10.47, 10.47,
10.46, 10.46, 10.46, 10.46, 10.46, 10.46, 10.45, 10.45, 10.45,
10.45, 10.45, 10.45, 10.44, 10.44, 10.44, 10.43, 10.43, 10.43,
10.43, 10.42, 10.42, 10.42, 10.42, 10.42, 10.41, 10.41, 10.4,
10.4, 10.4, 10.4, 10.4, 10.39, 10.39, 10.39, 10.38, 10.38, 10.37,
10.37, 10.37, 10.35, 10.35, 10.35, 10.34, 10.33, 10.33, 10.33,
10.32, 10.32, 10.3, 10.3, 10.3, 10.29, 10.29, 10.29, 10.29, 10.29,
10.28, 10.27, 10.27, 10.27, 10.27, 10.27, 10.26, 10.26, 10.26,
10.26, 10.25, 10.25, 10.25, 10.24, 10.24, 10.23, 10.23, 10.23,
10.22, 10.22, 10.22, 10.22, 10.21, 10.21, 10.21, 10.2, 10.19,
10.19, 10.19, 10.19, 10.18, 10.18, 10.18, 10.17, 10.17, 10.16,
10.16, 10.16, 10.16, 10.16, 10.15, 10.15, 10.13, 10.13, 10.13,
10.12, 10.11, 10.11, 10.09, 10.09, 10.08, 10.08, 10.07, 10.06,
10.05, 10.04, 10.04, 10.03, 10.03, 10.02, 10.02, 10.01, 9.999,
9.976, 9.967, 9.964, 9.955, 9.951, 9.939, 9.939, 9.895, 9.894,
9.888, 9.882, 9.858, 9.857, 9.815, 9.811, 9.809, 9.79, 9.759,
9.719, 9.718, 9.677, 9.674, 9.666, 9.651, 9.581, 9.567, 9.536,
9.508, 9.427, 9.385, 9.343, 9.254, 9.188, 9.03, 8.724), sample_type = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L,
2L, 1L, 2L), levels = c("Solid Tissue Normal", "Primary Tumor"
), class = "factor")), row.names = c(NA, -564L), class = "data.frame")
> dput(geneC)
structure(list(geneC = c(11.24, 11.14, 10.77, 10.75, 10.71, 10.66,
10.65, 10.65, 10.63, 10.62, 10.62, 10.61, 10.61, 10.59, 10.56,
10.55, 10.54, 10.53, 10.53, 10.51, 10.5, 10.47, 10.45, 10.44,
10.44, 10.42, 10.41, 10.4, 10.39, 10.38, 10.38, 10.38, 10.36,
10.35, 10.35, 10.35, 10.34, 10.34, 10.34, 10.33, 10.33, 10.33,
10.32, 10.31, 10.31, 10.3, 10.3, 10.3, 10.29, 10.27, 10.25, 10.24,
10.24, 10.23, 10.23, 10.23, 10.23, 10.22, 10.22, 10.22, 10.22,
10.21, 10.21, 10.21, 10.2, 10.2, 10.19, 10.19, 10.19, 10.19,
10.18, 10.17, 10.17, 10.17, 10.16, 10.16, 10.15, 10.15, 10.15,
10.15, 10.14, 10.13, 10.13, 10.13, 10.12, 10.11, 10.11, 10.11,
10.1, 10.1, 10.1, 10.09, 10.09, 10.09, 10.09, 10.08, 10.08, 10.08,
10.08, 10.08, 10.07, 10.07, 10.07, 10.06, 10.05, 10.05, 10.04,
10.04, 10.04, 10.03, 10.02, 10.02, 10.02, 10.02, 10.01, 10, 10,
10, 9.993, 9.993, 9.993, 9.991, 9.991, 9.989, 9.988, 9.984, 9.981,
9.981, 9.977, 9.975, 9.973, 9.973, 9.973, 9.972, 9.971, 9.97,
9.969, 9.966, 9.965, 9.962, 9.962, 9.96, 9.959, 9.958, 9.954,
9.946, 9.944, 9.943, 9.941, 9.937, 9.936, 9.935, 9.935, 9.932,
9.927, 9.925, 9.923, 9.919, 9.913, 9.91, 9.909, 9.908, 9.908,
9.906, 9.897, 9.896, 9.892, 9.889, 9.888, 9.888, 9.885, 9.885,
9.884, 9.883, 9.882, 9.874, 9.873, 9.873, 9.872, 9.868, 9.865,
9.858, 9.856, 9.845, 9.839, 9.835, 9.828, 9.82, 9.81, 9.805,
9.804, 9.804, 9.798, 9.788, 9.788, 9.787, 9.785, 9.785, 9.784,
9.783, 9.779, 9.778, 9.774, 9.773, 9.769, 9.768, 9.761, 9.747,
9.745, 9.745, 9.745, 9.743, 9.742, 9.733, 9.728, 9.728, 9.726,
9.718, 9.715, 9.714, 9.712, 9.71, 9.709, 9.709, 9.709, 9.703,
9.703, 9.696, 9.691, 9.688, 9.686, 9.682, 9.681, 9.677, 9.674,
9.669, 9.668, 9.663, 9.662, 9.657, 9.656, 9.648, 9.647, 9.645,
9.642, 9.642, 9.642, 9.636, 9.634, 9.63, 9.624, 9.618, 9.614,
9.614, 9.613, 9.613, 9.611, 9.611, 9.61, 9.595, 9.593, 9.59,
9.585, 9.584, 9.581, 9.58, 9.575, 9.575, 9.574, 9.571, 9.568,
9.565, 9.565, 9.564, 9.564, 9.561, 9.558, 9.555, 9.555, 9.554,
9.549, 9.546, 9.545, 9.541, 9.537, 9.532, 9.531, 9.53, 9.529,
9.528, 9.521, 9.521, 9.519, 9.519, 9.517, 9.516, 9.516, 9.514,
9.513, 9.512, 9.511, 9.51, 9.509, 9.508, 9.501, 9.5, 9.497, 9.494,
9.489, 9.489, 9.486, 9.483, 9.468, 9.463, 9.463, 9.458, 9.457,
9.454, 9.45, 9.443, 9.442, 9.442, 9.436, 9.432, 9.432, 9.431,
9.431, 9.429, 9.429, 9.428, 9.426, 9.426, 9.423, 9.423, 9.42,
9.418, 9.417, 9.41, 9.405, 9.405, 9.402, 9.399, 9.398, 9.395,
9.393, 9.392, 9.392, 9.39, 9.385, 9.383, 9.377, 9.37, 9.368,
9.367, 9.364, 9.361, 9.361, 9.36, 9.356, 9.349, 9.342, 9.342,
9.34, 9.339, 9.338, 9.331, 9.327, 9.326, 9.323, 9.319, 9.319,
9.312, 9.307, 9.304, 9.303, 9.3, 9.293, 9.292, 9.29, 9.289, 9.283,
9.271, 9.268, 9.263, 9.257, 9.256, 9.255, 9.255, 9.25, 9.25,
9.248, 9.246, 9.241, 9.24, 9.239, 9.239, 9.238, 9.237, 9.237,
9.211, 9.205, 9.203, 9.193, 9.193, 9.193, 9.188, 9.186, 9.182,
9.181, 9.177, 9.176, 9.173, 9.172, 9.159, 9.158, 9.158, 9.151,
9.146, 9.135, 9.134, 9.133, 9.133, 9.125, 9.123, 9.116, 9.114,
9.112, 9.112, 9.097, 9.092, 9.079, 9.079, 9.074, 9.064, 9.057,
9.053, 9.052, 9.049, 9.035, 9.031, 9.03, 9.026, 9.021, 9.02,
9.016, 9.012, 9.009, 9.008, 9.007, 8.996, 8.995, 8.991, 8.981,
8.975, 8.968, 8.965, 8.964, 8.963, 8.938, 8.929, 8.918, 8.918,
8.914, 8.913, 8.909, 8.908, 8.901, 8.897, 8.895, 8.892, 8.886,
8.886, 8.88, 8.872, 8.867, 8.866, 8.857, 8.854, 8.85, 8.848,
8.842, 8.835, 8.83, 8.829, 8.822, 8.814, 8.811, 8.808, 8.794,
8.792, 8.78, 8.777, 8.771, 8.761, 8.745, 8.745, 8.736, 8.731,
8.73, 8.728, 8.727, 8.717, 8.714, 8.713, 8.686, 8.68, 8.678,
8.645, 8.635, 8.614, 8.612, 8.592, 8.588, 8.587, 8.586, 8.58,
8.575, 8.571, 8.557, 8.549, 8.544, 8.511, 8.498, 8.485, 8.458,
8.458, 8.453, 8.451, 8.383, 8.347, 8.34, 8.338, 8.333, 8.308,
8.298, 8.275, 8.261, 8.249, 8.221, 8.212, 8.136, 8.134, 8.13,
8.093, 8.002, 8.001, 7.995, 7.981, 7.977, 7.97, 7.963, 7.946,
7.944, 7.938, 7.913, 7.844, 7.84, 7.58, 7.523, 7.518, 7.487,
7.414, 6.959, 6.212), sample_type = structure(c(2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L,
2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L
), levels = c("Solid Tissue Normal", "Primary Tumor"), class = "factor")), row.names = c(NA,
-564L), class = "data.frame")
CodePudding user response:
Did some data get filtered out? Try adding:
ylim(c('lower limit','upper limit'))
CodePudding user response:
The issue is that you did not take care of the sample type when cbinding the three datasets, e.g. for row 36 we have Solid Tissue Normal for geneA but Primary Tumor for B and C.
geneA[36,]
#> geneA sample_type
#> 36 7.822 Solid Tissue Normal
geneB[36,]
#> geneB sample_type
#> 36 11.38 Primary Tumor
geneC[36,]
#> geneC sample_type
#> 36 10.35 Primary Tumor
However, when you do cbind(geneA[,1],geneB[,1],geneC) all three genes are assigned the sample type from C, i.e. obs 36 for A is assigned Primary Tumor.
To fix that I would suggest to row bind your dataset using e.g. dplyr::bind_rows where as an intermediate step I first rename the gene columns. After doing so everything works fine and the there is no difference between the grouped plot and the separate plots:
library(dplyr, warn = FALSE)
library(ggplot2)
genes.df2 <- dplyr::lst(geneA, geneB, geneC) |>
lapply(rename_with, ~"Normalized_Counts", starts_with("gene")) |>
bind_rows(.id = "Gene")
ggplot(genes.df2, aes(x= Gene, y = Normalized_Counts, fill = sample_type))
geom_boxplot()
labs(title = "Gene Expression", x= "Gene", y = "Log2 normalized counts", fill = NULL)
theme(plot.title = element_text(hjust = 0.5))