Suppose this is the data at hand:
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
import seaborn as sns
data = {'trajectory': [101,102,102,102,102,102,102,102,104,104,104,104,104,104,104,107,107,107,107,
107,107,107,107,107,108,108,108,108,108,108,108,109,109,109,109,109,109,112,
112,112,112,112,113,113,113,113,114,114,114,114],
'segment': [1,1,1,1,2,2,3,3,1,1,2,2,2,3,3,1,1,2,2,2,2,3,3,3,1,1,1,
2,2,2,2,1,1,1,2,2,2,1,1,2,2,2,1,2,2,3,1,2,2,2],
'prediction': [3,0,0,1,3,3,2,2,0,0,4,4,2,0,0,0,0,2,2,2,3,0,0,2,0,0,1,1,
1,1,0,1,2,1,3,3,3,1,1,4,4,2,1,4,4,3,0,3,3,2]}
df = pd.DataFrame(data)
df.head(2)
trajectory segment prediction
0 101 1 3
1 102 1 0
And this is plotted like so:
plot_data = (df.value_counts()
.sort_values(ascending=False)
.reset_index()
.drop_duplicates(['trajectory', 'segment'])
.pivot_table(index='trajectory', columns='segment', values='prediction',))
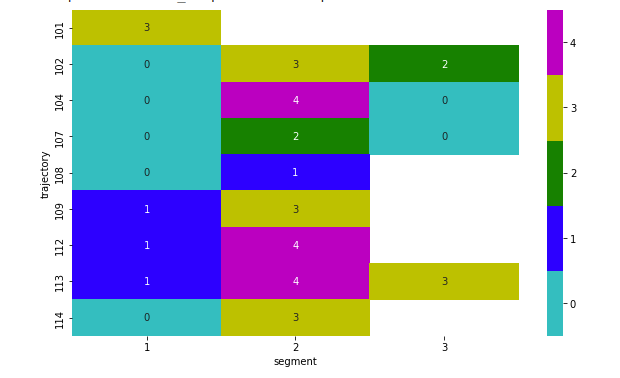
cmap = mcolors.ListedColormap(['c', 'b', 'g', 'y','m', ])
fig, ax = plt.subplots(figsize=(10,6))
sns.heatmap(plot_data,vmin=-0.5, vmax=4.5,cmap=cmap, annot=True)
Giving:
I want to fill all white cells to black. For that I have to replace all NaN values in my plot_data to some value, say 99, and add black color code k to cmap.
plot_data = (df.value_counts()
.sort_values(ascending=False)
.reset_index()
.drop_duplicates(['trajectory', 'segment'])
.pivot_table(index='trajectory', columns='segment', values='prediction',
fill_value=99))
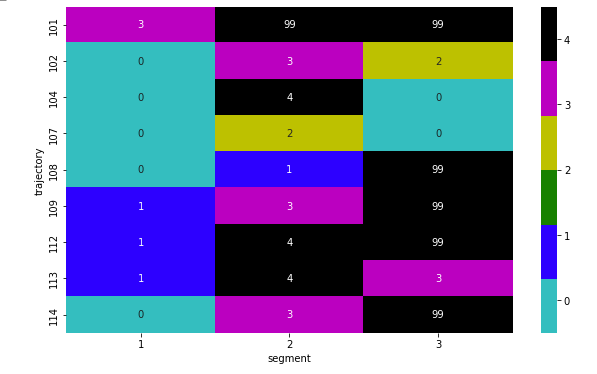
cmap = mcolors.ListedColormap(['c', 'b', 'g', 'y','m', 'k'])
fig, ax = plt.subplots(figsize=(10,6))
sns.heatmap(plot_data,vmin=-0.5, vmax=4.5,cmap=cmap, annot=True)
And plot again, giviing:
Confusion: 4 is coloured k: black, same as 99, instead of m: magenta. Plus, I do not like to annotate the null value cells with 99. It is there as a placeholder, since I cannot plot when NaN values are replaced with character such as -.
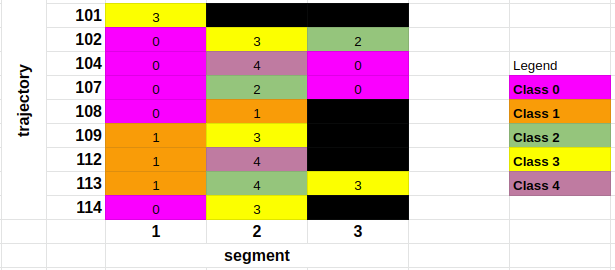
Intended results: something like the following
CodePudding user response:
You can use set_bad to set the color for masked values of your colorbar to opaque black:
cmap = mcolors.ListedColormap(['c', 'b', 'g', 'y','m',])
cmap.set_bad('k')
(in your colormap definition it's transparent black, that's why you can see the Axes patch in the first place).
CodePudding user response:
Ah, All I need was to set the background to black, before adding heatmap, like so:
ax.set_facecolor('black')
sns.heatmap(plot_data, vmin=-0.5, vmax=4.5, cmap=cmap, annot=True)
And that's it.