I am trying to understand why my code produces a different result when run with reprex::reprex() than directly from the script and how to consistently produce the output of the reprex() call. The issue emerges within the filter() call.
- Example 1 shows my function filters the data.frame rows based on a column's matches with another vector when I select, copy, and then run it with
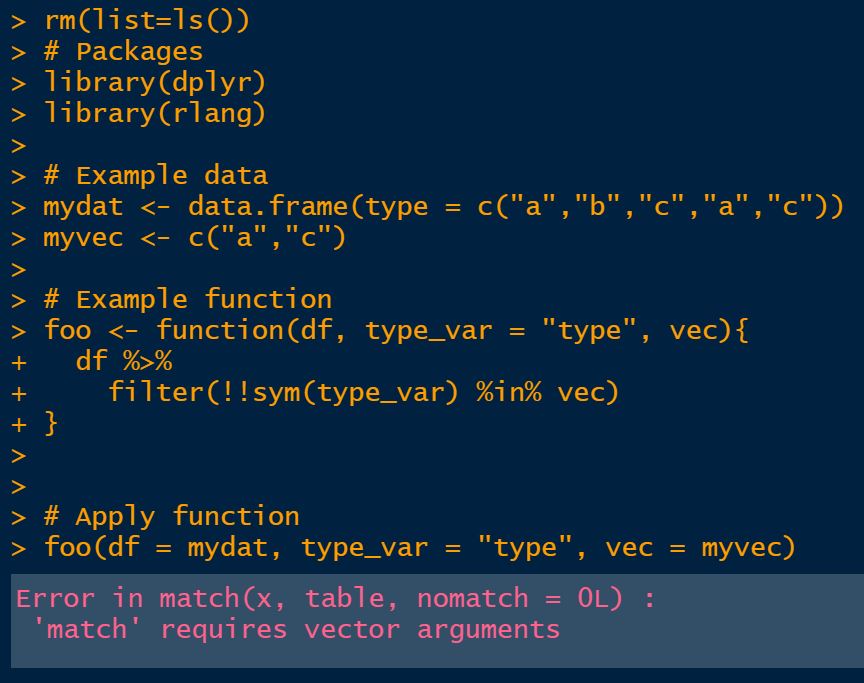
reprex::reprex()in RStudio. - Example 2 (screenshot from the console output) shows that running the exact same code directly in the script throws a
'match' requires vector argumentserror. - Example 3 shows with a slight modification of the function that
!!sym()appears to be creating some sort of time series object. Omittingsym()and replace==with%in%has the same consequence.
UPDATE:
The issue did not replicate on others' machines nor my own. I swapped out of an RStudio project to a single .R file and it still persisted. However, when I Cntrl Shift F10 to detach libraries, data, etc. the discrepancy vanished. This suggested that I was deal with some sort of namespace issue. Upon returning to the RStudio Project, the issue returned. However, calling dplyr::filter() within the function resolved the issue - reinforcing it being a namespace issue.
While the accepted answer provides some solutions and correctly identifies the issue, the outstanding question (for another post) is why the namespace precedence was not applied in this case when I loaded the package immediately beforehand.
Example 1: !!sym() produces a vector for %in% as expected when code is run with reprex::reprex()
# Packages
library(dplyr)
library(rlang)
# Example data
mydat <- data.frame(type = c("a","b","c","a","c"))
myvec <- c("a","c")
# Example function
foo <- function(df, type_var = "type", vec){
df %>%
filter(!!sym(type_var) %in% vec)
}
# Call function
foo(df = mydat, type_var = "type", vec = myvec)
#> type
#> 1 a
#> 2 c
#> 3 a
#> 4 c
Example 2: Console output shows type error when run from within an R script
Example 3: slightly modified function shows that !!sym() is creating a time series object?!
# Example function
foo <- function(df, type_var = "type", vec){
df %>%
filter(!!sym(type_var) == "a")
}
# Apply function
foo(df = mydat, type_var = "type", vec = myvec)
#>Time Series:
#>Start = 1
#>End = 5
#>Frequency = 1
#> [,1]
#> [1,] 0
#> [2,] 0
#> [3,] 0
#> [4,] 0
#> [5,] 0
CodePudding user response:
It's related to which version of filter is being used and whether it's imported from stats or dplyr. I suspect you have an ~/.Rprofile somewhere that's loading some library functions which are being loaded sometimes and not others.
Changing example 3 to
foo <- function(df, type_var = "type", vec){
df %>%
dplyr::filter(!!sym(type_var) == "a")
}
# Apply function
foo(df = mydat, type_var = "type", vec = myvec)
yields:
type
1 a
2 a
Similarly changing example 1 to:
library(dplyr)
library(rlang)
# Example data
mydat <- data.frame(type = c("a","b","c","a","c"))
myvec <- c("a","c")
# Example function
foo <- function(df, type_var = "type", vec){
df %>%
dplyr::filter(!!sym(type_var) %in% vec)
}
# Call function
foo(df = mydat, type_var = "type", vec = myvec)
gives:
type
1 a
2 c
3 a
4 c
Beware of namespace collisions when running R in console/Rscript etc, it can be hard to track down bugs. filter and lag are the chief culprits (source I almost had to retract a journal paper because lag was imported from the wrong namespace on an Rscript and failed in a weird and silent way).