I am using the survival package to make Kaplan-Mayer estimates of survival curves by group and then I plot out the said curves using packages ggfortify and survminer. All works fine except the legend labels for plotting. I want to present N sizes of groups in the legend labels. I thought that adding the N size to the grouping variable itself using paste0 was a good way to go. In my case it is easier than to use something like scale_fill_discrete("", labels = legend_labeller_for_plot).
library(dplyr)
library(ggplot2)
library(survival)
library(survminer)
library(ggfortify)
set.seed = 100
data <- data.frame(
time = rlnorm(20),
event = as.integer(runif(20) < 0.5),
group = ifelse(runif(20) > 0.5,
"group A",
"group B")
)
# Plotting survival curves without N sizes in the legend
fit <- survfit(
with(data, Surv(time, event)) ~ group,
data)
autoplot(fit)
# Adding N sizes to the data and plotting
data_new <- data %>%
group_by(group) %>% mutate(N = n()) %>%
ungroup() %>%
mutate(group_with_N = paste0(group, ", N = ", N))
fit_new <- survfit(
with(data, Surv(time, event)) ~ group_with_N,
data_new)
autoplot(fit_new)
When I try to add N sizes to the groups variable, the part with "N =" in the grouping variable disappears, i.e. the group variable isn't displayed on the legend labels as expected.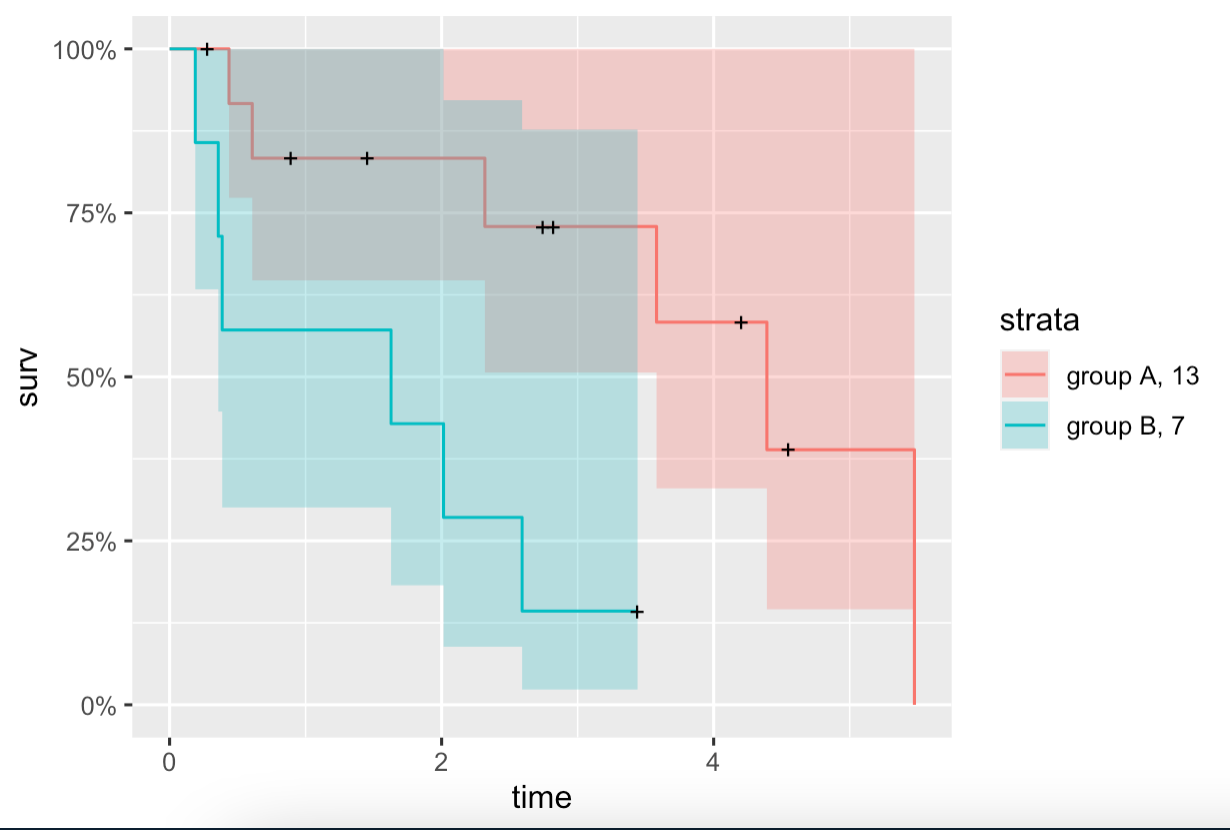
For comparison, what I expect is something like the following using Iris data:
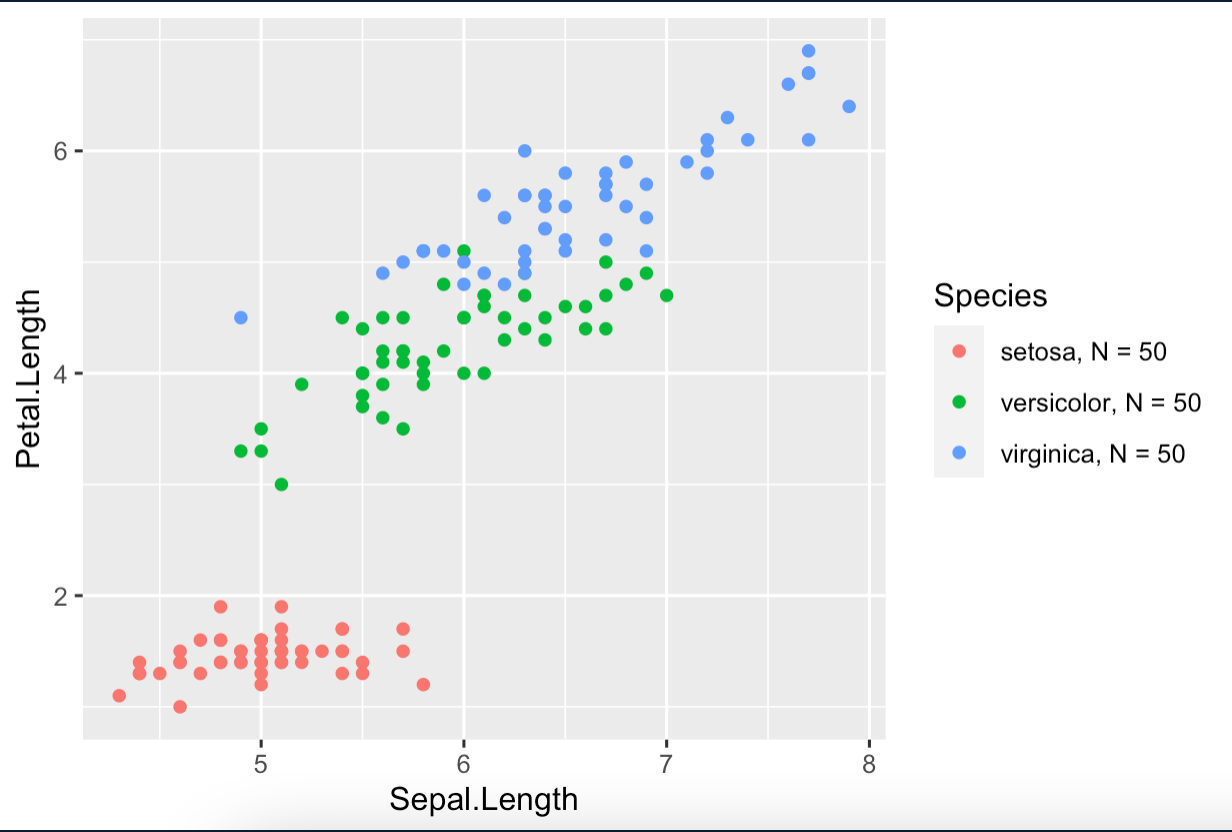
What is more, I found that that the culprit is the equali sign =. When I remove the = sign, the legend labels correspond to the grouping variable values.
My question is, why does the equal sign cause this?
CodePudding user response:
An option could be using ggsurvplot where you can specify the legend.labs so you can show your size in the legend like this:
library(dplyr)
library(ggplot2)
library(survival)
library(survminer)
library(ggfortify)
set.seed = 100
data <- data.frame(
time = rlnorm(20),
event = as.integer(runif(20) < 0.5),
group = ifelse(runif(20) > 0.5,
"group A",
"group B")
)
# Adding N sizes to the data and plotting
data_new <- data %>%
group_by(group) %>% mutate(N = n()) %>%
ungroup() %>%
mutate(group_with_N = paste0(group, ", N = ", N))
fit_new <- survfit(
with(data, Surv(time, event)) ~ group_with_N,
data_new)
p <- autoplot(fit_new)
p
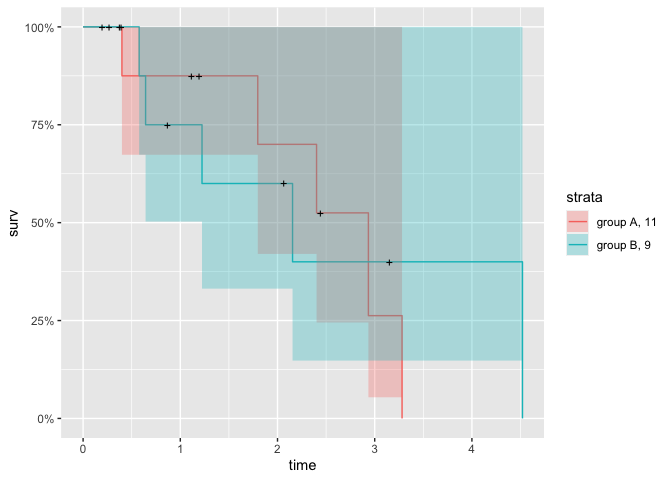
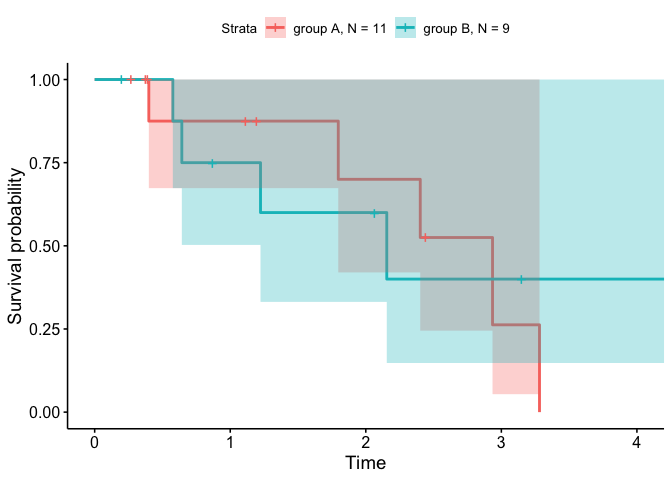
# ggsurvplot
ggsurvplot(fit_new, data_new,
legend.labs = unique(sort(data_new$group_with_N)),
conf.int = TRUE)
Created on 2022-08-18 with reprex v2.0.2
