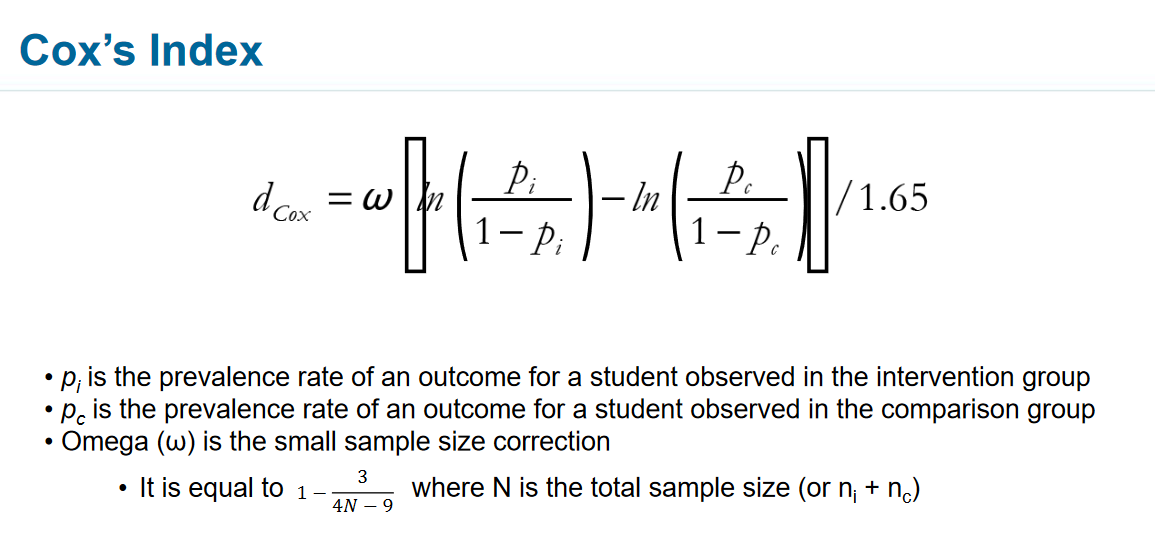
I would like to find cox's index as indicated by the formula below.
Here is an example of my data;
pass_fl <- c(rep('1',5),rep('0',3), rep('0',2))
in_pass <- c(rep('1',4),rep('0',1),rep('1',5))
treatment_fl <- c(rep('0',2),rep('1',3),rep('1',2),rep('0',3))
df <- data.frame(pass_fl, treatment_fl, in_pass) %>%
mutate_all(as.numeric)
Where pass_fl(p) is the outcome I am interested in, and in_pass indicates whether each record is should be included in the calculation.
- Is there an existing R package/function I could use?
- If not, what is the best way of setting up this formula?
This is what I tried:
library(SciViews) First, subset to those in the sample only?
df_pass <- df %>% filter(in_pass==1)
So that N = 3 3??
Omega (ω) is the small sample size correction
omega = 1 - (3/(4*6-9))
where total sample size = 6
pi is the prevalence rate of an outcome for a student observed in the intervention group
pi_dat <- df_pass[which(df_pass$treatment_fl==1),]
pi <- mean(pi_dat$pass_fl, na.rm = T)
pi
pc is is the prevalence rate of an outcome for a student observed in the comparison group
pc_dat <- df_pass[which(df_pass$treatment_fl==0),]
pc <- mean(pc_dat$pass_fl,na.rm = T)
pc
cox index
cox = omega * abs(ln(pi/1-pi) -abs(ln(pc/1-pc)))/1.65
cox
Thanks for your help!
CodePudding user response:
I don't have enough reputation to comment, but could you be slightly more specific about how the columns in your example data correspond to the variables in your formula? Below is my guess, which currently returns Inf for the example data.
library(data.table)
setDT(df)
N <- df[in_pass == 1, .N]
omega <- 1 - (3/(4*N - 9))
p_i <- df[in_pass == 1][treatment_fl == 1, sum(pass_fl == 1)/.N]
p_c <- df[in_pass == 1][treatment_fl == 0, sum(pass_fl == 1)/.N]
d_cox <- omega * abs(log(p_i/(1-p_i)) - log(p_c/(1-p_c))) / 1.65 # Inf