Here is my data:
df1=
structure(list(time = structure(c(-2209007520, -2209009560, -2209034520,
-2209019460, -2209035180, -2209028880, -2209011480, -2209040100,
-2209013340, -2208991200, -2209038540, -2209030080, -2209027380,
-2209014120, -2209034280, -2209019460, -2208994020, -2209051980,
-2209038480, -2209026600, -2209013580, -2209037220, -2209029420,
-2209022100, -2208999120, -2209012560, -2209040280, -2209029300,
-2209027620, -2209025400, -2209063920, -2209044480, -2209022520,
-2209065000, -2209052940, -2209004880, -2208989940, -2209074960,
-2209053480, -2208997680, -2209069500, -2209069440, -2209061580,
-2209023300, -2209029600, -2209039380, -2209030020, -2209016700,
-2209016460, -2209035660, -2209030020, -2209067520, -2209016880,
-2209074300, -2209029660, -2209020780, -2208991500, -2209043760,
-2209013340, -2208997920, -2208990120, -2209017600, -2209041420,
-2209040280, -2209040400, -2209066080, -2209072440, -2209034340,
-2209049760, -2209051560, -2209031700, -2209035720, -2209069260,
-2209048800, -2209036980, -2209037520, -2209038540, -2209050900,
-2208998160, -2209029120, -2209013040, -2209056240, -2209027260,
-2209012260, -2209014360, -2209007220, -2209003440), tzone = "UTC", class = c("POSIXct",
"POSIXt")), result = c(7.6, 18, 10.1, 9.3, 19, 14.4, 14.9, 11.2,
16.1, 10.9, 8.2, 15.1, 9.4, 13.2, 17.5, 8.4, 7.2, 19, 14, 19.1,
12.4, 11.9, 16.2, 19.1, 18.6, 15.1, 13.1, 5.6, 18, 18.7, 19.4,
20.4, 10, 20.9, 24.4, 12.5, 10.7, 11.6, 6.8, 5.2, 23.4, 9.4,
25.6, 13.7, 22.1, 20.2, 15.5, 17.1, 18.5, 18.7, 19.1, 21.8, 13.9,
4.5, 17.8, 2.6, 22.5, 20.6, 9.6, 6.5, 7.1, 13.2, 22.7, 14.2,
21.8, 19.9, 18.8, 10.1, 9.1, 15.5, 12.2, 12.5, 13.7, 12.7, 13.3,
14.8, 13.3, 16.1, 20.9, 15.8, 20, 14.1, 21.6, 19.2, 24, 19.8,
16.6)), row.names = c(NA, -87L), class = c("tbl_df", "tbl", "data.frame"
))
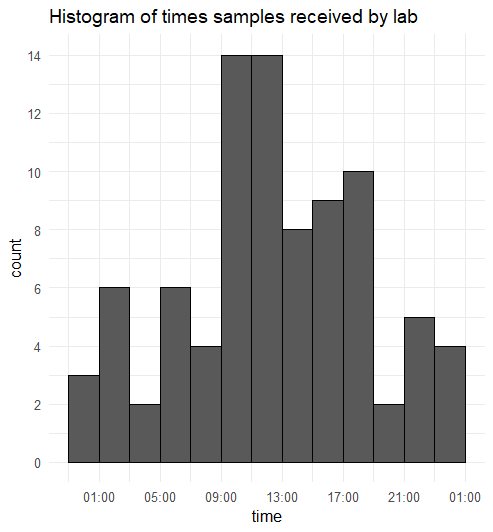
The date in the time column is not the true date. I'm only interested in the times. I was going to convert these into periods with lubridate::hms() but was struggling to plot that.
I have created this histogram
ggplot(df1, aes(x= time))
geom_histogram(binwidth= 7200, colour = "black")
scale_x_datetime(date_breaks = "4 hour", date_labels = "%H:00")
scale_y_continuous(breaks = seq(0,30,2))
theme_minimal()
labs(title = "Histogram of times samples received by lab")
Why has the x-axis extended beyond 24 hours despite all times being on the same date? How can I add the two 23:00 to 01:00 bars together?
CodePudding user response:
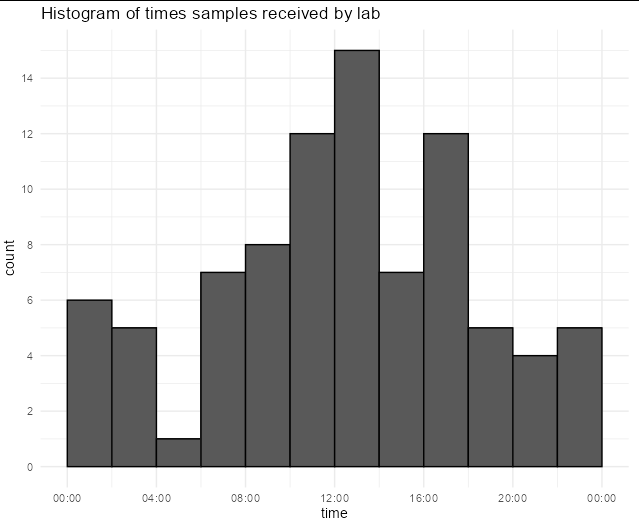
You can specify the histogram breaks as well as the x axis breaks in terms of POSIXct date-times to get full control over the x axis:
ggplot(df1, aes(x= time))
geom_histogram(breaks = seq(as.POSIXct('1899-12-31 00:00:00'),
as.POSIXct('1900-01-01 00:00:00'), "7200 sec"),
colour = "black")
scale_x_datetime(breaks = seq(as.POSIXct('1899-12-31 00:00:00'),
as.POSIXct('1900-01-01 00:00:00'), "14400 sec"),
date_labels = "%H:00")
scale_y_continuous(breaks = seq(0,30,2))
theme_minimal()
labs(title = "Histogram of times samples received by lab")