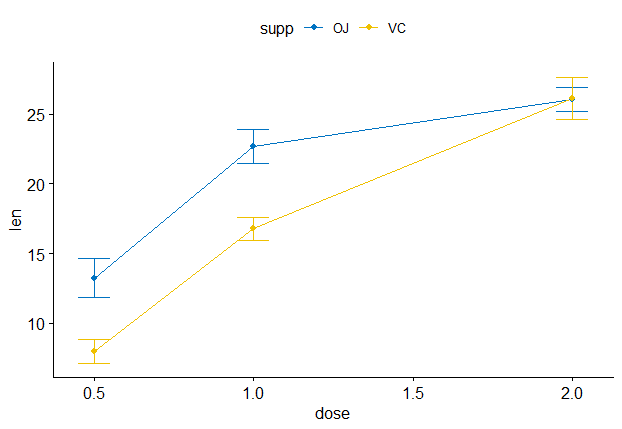
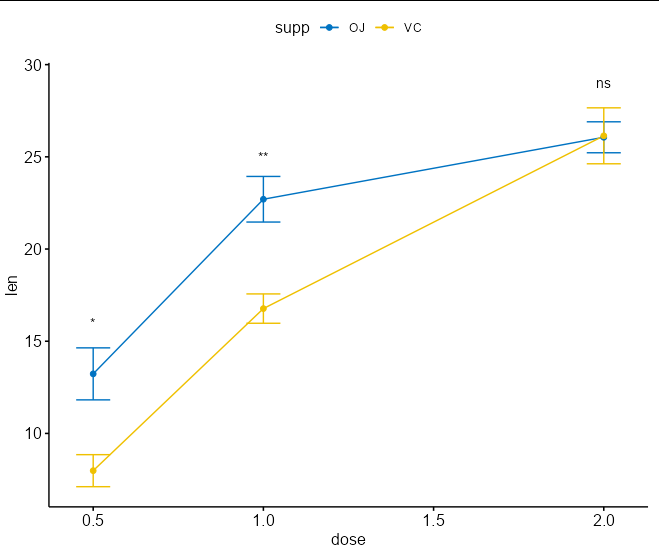
I am trying to have a numeric axis in R, ggline with statistical test. In my data instead of dose, I have time on my x axis.
Without continuous scale it works, as described (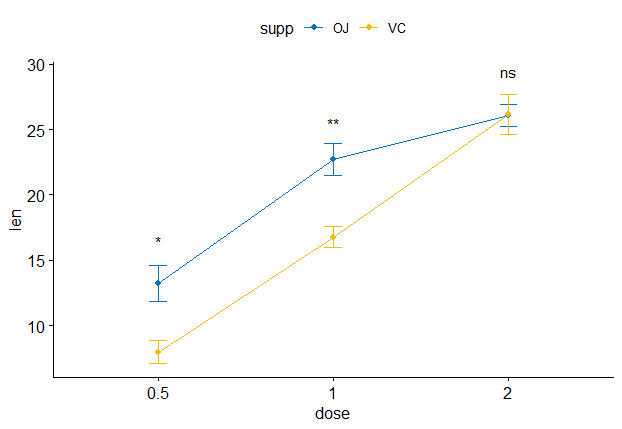
Changing the axis to look how I want (continuous/numeric for numbers) using this code, the stat_compare_means is not working anymore:
ggline(ToothGrowth, x = "dose", y = "len", add = "mean_se",
color = "supp", palette = "jco", numeric.x.axis = TRUE)
stat_compare_means(aes(group = supp), label = "p.signif",
label.y = c(16, 25, 29))
I get the following error message:
Computation failed in
stat_compare_means(): '*.npc coord for x axis should be either a numeric value in [0-1] or a character strings including one of right, left, center, centre, middle
My aim is to get the axis as in the second plot, but to include the statistics from the first plot. It would be great if anyone knows how to fix this (:
CodePudding user response:
This doesn't seem possible directly within stat_compare_means. However, you can get the same backend function, compare_means, to create the data required to draw the labels using geom_text:
library(tidyverse)
library(ggpubr)
data("ToothGrowth")
ggline(ToothGrowth, x = "dose", y = "len", add = "mean_se",
color = "supp", palette = "jco", numeric.x.axis = TRUE)
geom_text(aes(x = dose, y = c(16, 25, 29), label = p.signif),
color = "black", inherit.aes = FALSE,
data = ToothGrowth %>%
nest(data = -dose) %>%
mutate(data2 = map(data, ~ compare_means(len ~ supp, .x,
ref.group = "VC"))) %>%
unnest(data2))