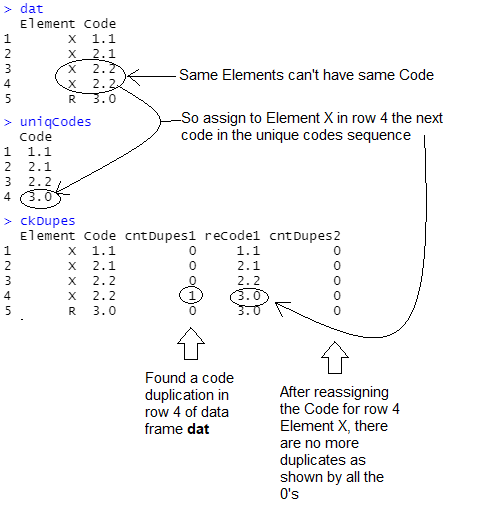
I'm working on looping code in R that identifies any disallowed duplications in a sequence of numbers, and then reiteratively recasts the numbers into a correct sequence. As shown in the image below starting with the first step in an assumed loop, where dat is the initial data frame whose sequences are to be checked, uniqCodes isolates the unique Codes from dat, and then ckDupes counts any disallowed duplicates in column cntDupes1, reassigns the next sequential Code in those instances of disallowed duplicates in column reCode1, and then another check for disallowed duplicates in column cntDupes2:
Below is my code for these 3 data frames, that works in this scenario. Is there a more efficient way to do the duplication identification and resequencing currently done in ckDupes? This is the kind of tangled code that I won't understand a month from now and it needs to be clear and efficient since it'll be in an iterative process as I build out the loop.
Code:
library(dplyr)
dat <- data.frame(Element= c("X","X","X","X","R"),
Code = c(1.1,2.1,2.2,2.2,3))
uniqCodes <- dat %>% select(Code) %>% distinct()
ckDupes <- dat %>%
mutate(cntDupes1 = sapply(1:n(), function(x) sum(Element[1:x]==Element[x] & Code[1:x] == Code[x]))-1) %>%
mutate(reCode1 = ifelse(cntDupes1 > 0,uniqCodes[match(Code,uniqCodes$Code) cntDupes1,],Code)) %>%
mutate(cntDupes2 = sapply(1:n(), function(x) sum(Element[1:x]==Element[x] & reCode1[1:x] == reCode1[x]))-1)
CodePudding user response:
Let me give a few options, at least they prevent to loop and recheck for new duplicates over and over again. Lets take this little expanded dataset.
dat <- data.frame(Element= c("X", "X", "X","X","X", "X","R", "Z", "Z", "Z"),
Code = c(1.1, 1.1, 1.1, 1.2, 2.0, 3, 3, 10, 4.4, 4.5))
Using your code first
uniqCodes <- dat %>% select(Code) %>% distinct()
dat %>%
mutate(cntDupes1 = sapply(1:n(), function(x) sum(Element[1:x]==Element[x] & Code[1:x] == Code[x]))-1) %>%
mutate(reCode1 = ifelse(cntDupes1 > 0,uniqCodes[match(Code,uniqCodes$Code) cntDupes1,],Code)) %>%
mutate(cntDupes2 = sapply(1:n(), function(x) sum(Element[1:x]==Element[x] & reCode1[1:x] == reCode1[x]))-1)
# Element Code cntDupes1 reCode1 cntDupes2
# 1 X 1.1 0 1.1 0
# 2 X 1.1 1 1.2 0
# 3 X 1.1 2 2.0 0
# 4 X 1.2 0 1.2 1
# 5 X 2.0 0 2.0 1
# 6 X 3.0 0 3.0 0
# 7 R 3.0 0 3.0 0
# 8 Z 10.0 0 10.0 0
# 9 Z 4.4 0 4.4 0
# 10 Z 4.5 0 4.5 0
Here we already see two serious issues in reCode1, you renamed the second 1.1 value to 1.2 while that already existed, then - imo not consistent - rename the 3rd 1.1 value to 2.0 which also existed.
Not sure on what conditions you want to rename or keep existing values or if it is not relevent at all as long they get a new unique code.
I use data.table below in my example with three variations to make them all unique. And here the three alternative Recoding suggestions.
- RecodedAlt1 - we keep the unique ones per Element "as is" and we replace the duplicates with a new number starting from the highest value (after the digit) in the group.
- We totally recode everything, starting with *.1 (RecodedAlt2) or *.0 (RecodedAlt3)
data.table
library(data.table)
setDT(dat)
dat[, dup := duplicated(Code), Element]
dat[, c("x", "y") := tstrsplit(sprintf("%.1f", Code), "\\.", type.convert = T), by = Element]
setorder(dat, x, y)
dat[, max := max(Code), by = .(Element, x)]
dat[dup == T, RecodedAlt1 := (1:.N)/ 10 max, by = .(Element, x)]
dat[dup == F, RecodedAlt1 := Code]
dat[, reCodedAlt2 := paste(x, 1:.N, sep = "."), by = .(Element, x)]
dat[, reCodedAlt3 := paste(x, 0:(.N-1), sep = "."), by = .(Element, x)]
results
dat
Element Code dup x y max RecodedAlt1 reCodedAlt2 reCodedAlt3
1: X 1.1 FALSE 1 1 1.2 1.1 1.1 1.0
2: X 1.1 TRUE 1 1 1.2 1.3 1.2 1.1
3: X 1.1 TRUE 1 1 1.2 1.4 1.3 1.2
4: X 1.2 FALSE 1 2 1.2 1.2 1.4 1.3
5: X 2.0 FALSE 2 0 2.0 2.0 2.1 2.0
6: X 3.0 FALSE 3 0 3.0 3.0 3.1 3.0
7: R 3.0 FALSE 3 0 3.0 3.0 3.1 3.0
8: Z 4.4 FALSE 4 4 4.5 4.4 4.1 4.0
9: Z 4.5 FALSE 4 5 4.5 4.5 4.2 4.1
10: Z 10.0 FALSE 10 0 10.0 10.0 10.1 10.0