I am trying to measure average thickness of segmented and labeled image. Since I was not successful to make this work with opencv, per suggestion by @CrisLuengo, I switch to using diplib. I found a good example of measuring thickness of part here 
Here I am interested in 3 average thicknesses:
- thickness of long narrow white part on top
- thickness of black region in middle
- thickness of thick white part in bottom
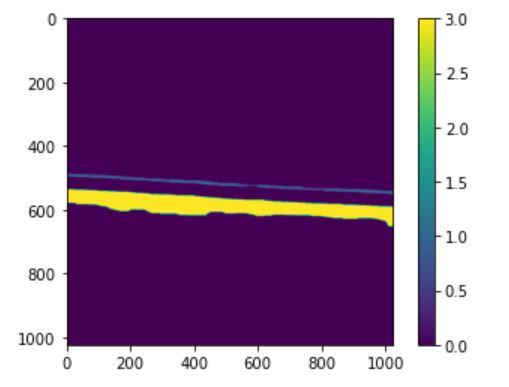
in order to do this first I labeled the image and chose top 2 big areas (which is top and bottom white part of image)
label_image=measure.label(opening1, connectivity=opening1.ndim)
props= measure.regionprops_table (label_image, properties=['label', "area", "coords"])
slc=label_image
rps=regionprops(slc)
areas=[r.area for r in rps]
id=np.argsort(props["area"])[::-1]
new_slc=np.zeros_like(slc)
for i in id[0:2]:
new_slc[tuple(rps[i].coords.T)]=i 1
which will result in labeled image with 2 labels:
Since, it looks like that the approach introduced here 
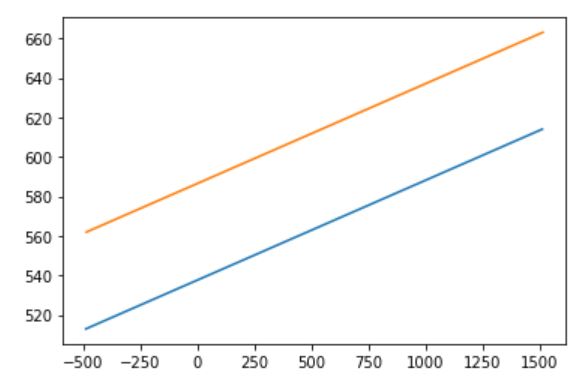
then I ran the code and it return 15.33 μm thickness (compared to the actual thickness of 14.66 μm this is a good estimation). it also returned this image
I am not sure how to interpret this image but it looks like the algorithm is trying to show fitting lines to boundaries now I want to do the same thing for thin white top part. To do this first I choose the top white part:
rps=regionprops(slc)
areas=[r.area for r in rps]
id=np.argsort(props["area"])[::-1]
new_slc=np.zeros_like(slc)
for i in id[1:2]:
new_slc[tuple(rps[i].coords.T)]=i 1
then I ran the algorithm explained here:
Can someone advise, why the algorithm is not working on the second portion of my image? what's the difference and why I am getting errors?
UPDATE regarding Pre-processing
====================
median=cv2.medianBlur(img,13)
ret, th = cv2.threshold(median, 0 , 255, cv2.THRESH_BINARY cv2.THRESH_OTSU)
kernel=np.ones((3,15),np.uint8)
closing1 = cv2.morphologyEx(th, cv2.MORPH_CLOSE, kernel, iterations=2)
kernel=np.ones((1,31),np.uint8)
closing2 = cv2.morphologyEx(closing1, cv2.MORPH_CLOSE, kernel)
label_image=measure.label(closing2, connectivity=closing2.ndim)
props= measure.regionprops_table (label_image, properties=['label'])
kernel=np.ones((1,13),np.uint8)
opening1= cv2.morphologyEx(closing2, cv2.MORPH_OPEN, kernel, iterations=2)
label_image=measure.label(opening1, connectivity=opening1.ndim)
props= measure.regionprops_table (label_image, properties=['label', "area", "coords"])
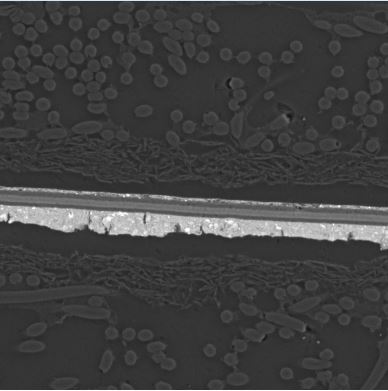
Original photo: Below I showed low quality of my original image, so you have better understanding as why I did all the pre-processing
CodePudding user response:
For the thin line the program from the other Q&A doesn't work because the two edges along the line are too close together, the program doesn't manage to identify the two as separate objects.
For a very thin line like this you could do something that is not possible with a thicker line: just measure its length and area, the width will be the ratio of those two:
# `label_image` is as in the OP
# `id` is the label ID for the thin line
msr = dip.MeasurementTool.Measure(label_image.astype(np.uint32), features=['Size','Feret'])
area = msr[id]['Size'][0]
length = msr[id]['Feret'][0]
width = area / length
Note that you should be able to get a more precise value if you don't immediately binarize the image. The linked Q&A uses grayscale input image to determine the location of the edges more precisely than you can do after binarizing.