I tried doing this
fits = list(fit0)
for(i in 1:5)
{
temp = assign(paste0("fit", i), lm(formula = y ~ poly(x, degree = i, raw = TRUE)))
fits = append(fits, temp)
}
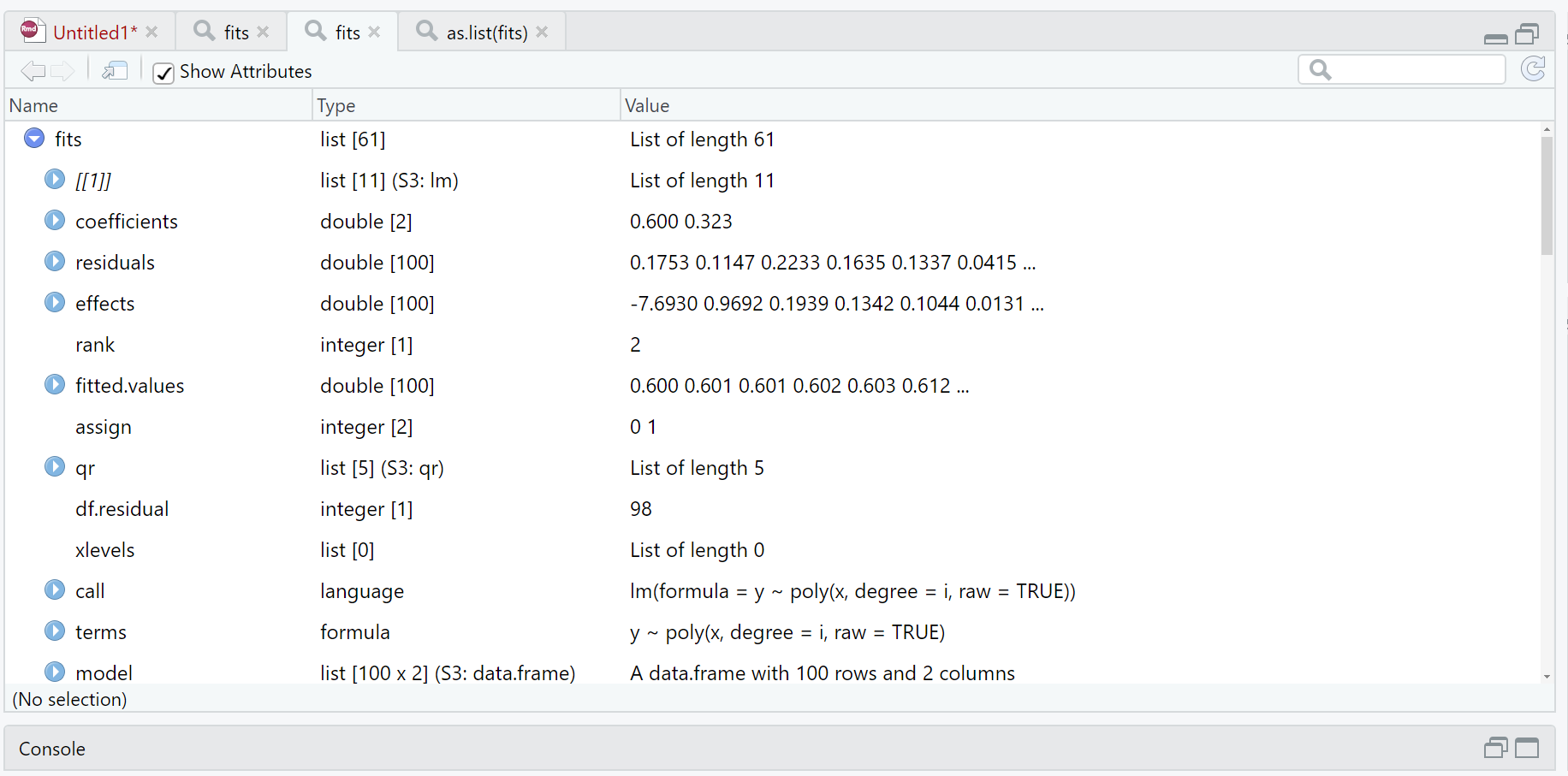
which seems like it should work and I don't get any errors initially. The problem though, is that instead of creating a list of lists of length 6, where each element is itself a list (as lm objects are lists), it seems to be taking the elements of each list and making them each a separate element in temp. When I do length(fits) it gives 61. And when I do View(fits) it shows me this:
which certainly looks like it took all the elements of each individual list and made them the elements of fits, though I don't understand why.
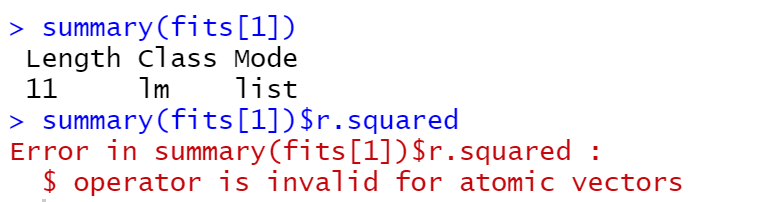
Oddly though, if I just do fits[1] in the console it gives
which is the exact same output I get if I type fit0 in the console. So it seems like it's in someway storing each lm object as one thing.
The problem though, is that if I then try to get the R^2 value for, e.g., fit0, it works fine if I do summary(fit0)$r.squared, but if try to do it for fits[1] it does this:
I don't understand what's going on here. I thought maybe the problem was using append, since I'd previously only used it with vectors so I Googled "how to create list of lists in R" but the examples I found used append so that doesn't seem to be the issue.
I assume it's something to do with the intricacy of lm objects, but the documentation isn't actually helpful (on a side note, why IS R's documentation so terrible anyway? Compared to Python, or even C (which is a far more complicated language to work with overall), it's so much harder to gleam the details of how the different functions and data types work because the documentation always seems to give the bear minimum, if that, of information) so I don't know what I'm doing wrong.
I've tried Googling how to create a list of lm objects and I found the lmlist data type documentation, but that seems to be for when you want to create a single regression but using data grouped by categories in a data.frame, which isn't what I'm trying to do here. I also found this post: 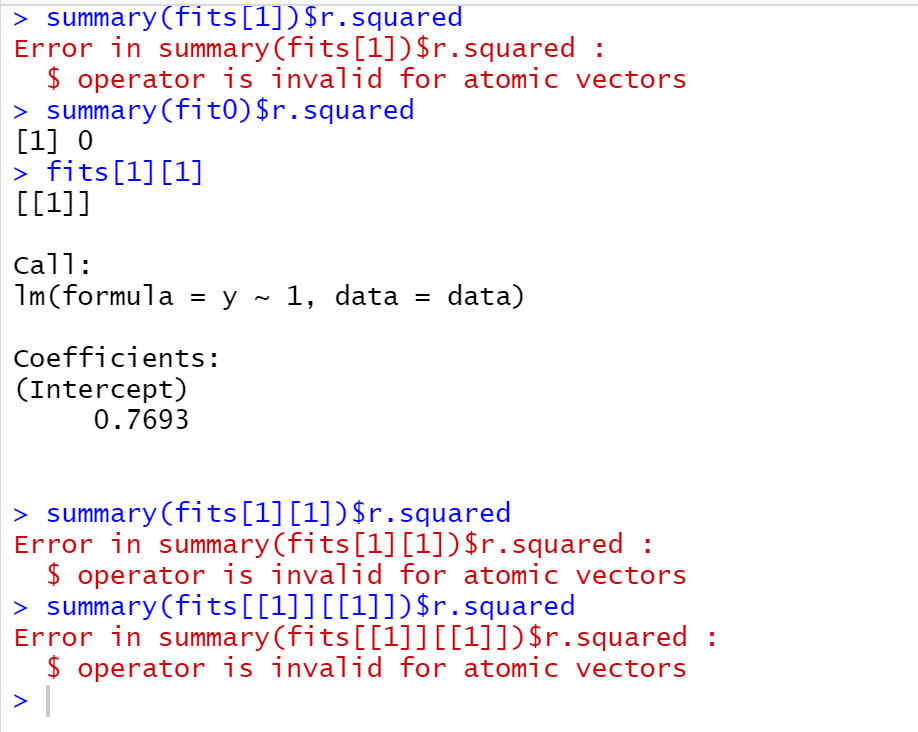
I'm quite confused here, so any guidance would be greatly appreciated.
CodePudding user response:
Showing how to use a for loop for this:
DF <- data.frame(x = rnorm(100), y = rnorm(100))
fits <- list(fit0 = lm(y ~ 1, data = DF))
for(i in 1:5)
{
fits[[paste0("fit", i)]] <- lm(formula = y ~ poly(x, degree = i, raw = TRUE), data = DF)
}
sapply(fits, \(x) summary(x)$r.squared)
# fit0 fit1 fit2 fit3 fit4 fit5
#0.00000000 0.06441347 0.07915820 0.08547018 0.08547089 0.08569820
From the perspective of a statistician, you should not do this.
CodePudding user response:
lm objects in R are indeed complicated. The broom package offers a consistent way to convert model objects into a "tidy" output format that can be easier to work with downstream.
For instance, we can use broom::glance to get a table with the lm stats as a data frame:
fit <- lm(mpg ~ wt, data = mtcars)
broom::glance(fit)
Result
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 0.753 0.745 3.05 91.4 1.29e-10 1 -80.0 166. 170. 278. 30 32
We could extend this to an example where we group the mtcars dataset by gear, nest the associated data for each gear group, run lm on each one, glance each of those, and finally unnest to get the results into a table. That seems to demonstrate what you're describing -- we can see how the r.squared varied for the lm run on each group.
library(tidyverse); library(broom)
mtcars %>%
group_by(gear) %>%
nest() %>%
mutate(fit = map(data, ~lm(.x$mpg~.x$wt)),
tidy = map(fit, glance)) %>%
unnest(tidy)
# Groups: gear [3]
gear data fit r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <list> <list> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 4 <tibble [12 × 10]> <lm> 0.677 0.645 3.14 21.0 0.00101 1 -29.7 65.4 66.8 98.9 10 12
2 3 <tibble [15 × 10]> <lm> 0.608 0.578 2.19 20.2 0.000605 1 -32.0 69.9 72.1 62.3 13 15
3 5 <tibble [5 × 10]> <lm> 0.979 0.972 1.11 141. 0.00128 1 -6.34 18.7 17.5 3.69 3 5
Or maybe you have your list of lm objects, you could feed those into map_dfr(glance) to get a table with r.squared:
fit1 <- lm(mpg~wt, mtcars)
fit2 <- lm(mpg~cyl wt, mtcars)
list(fit1, fit2) %>%
map_dfr(glance)
# A tibble: 2 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 0.753 0.745 3.05 91.4 1.29e-10 1 -80.0 166. 170. 278. 30 32
2 0.830 0.819 2.57 70.9 6.81e-12 2 -74.0 156. 162. 191. 29 32