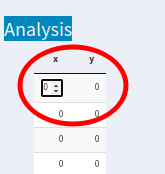
I'm trying to edit my datatable to put values as inputs. However, I would like to remove the numbers and scrolls from each cell, something similar to Excel/ SPSS cells. The problem is in the following image:
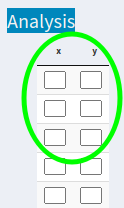
I would like all cells to look approximately like this as if they were Excel/ SPSS cells:
Table code:
library(shiny)
library(shinydashboard)
library(DT)
library(tidyverse)
library(shinyjs)
header <- dashboardHeader(title = "Dashboard", titleWidth = 300)
sidebar <- dashboardSidebar(
width = 300,
sidebarMenu(
menuItem(
text = "Menu 1",
tabName = "menu1",
icon = icon("chart-line")
)
)
)
body <- dashboardBody(
HTML(
"<head>
<script>
$(function() {
setTimeout(function() {
$('.dt-rigth').dblclick();
}, 1000);
});
</script>
</head>"
),
tabItems(
tabItem(
tabName = "menu1",
titlePanel(
title = HTML(
"<text style='background-color:#008cba; color:#f2f2f2;'>Analysis</text>"
)
),
fluidPage(
column(
id = "menusss1",
width = 12,
column(
id = "correl",
width = 1,
DT::DTOutput("my_datatable"),
actionButton("go", label = "Plot Data")
),
column(
id = "correlplot",
width = 6,
plotOutput("my_plot")
)
)
)
)
)
)
ui <- dashboardPage(header, sidebar, body)
server <- function(input, output) {
#initialize a blank dataframe
v <- reactiveValues(data = {
data.frame(x = numeric(0) ,y = numeric(0)) %>%
add_row(x = rep(0, 30) ,y = rep(0, 30))
})
#output the datatable based on the dataframe (and make it editable)
output$my_datatable <- DT::renderDataTable({
js <- "table.on('click', 'td', function() {
$(this).dblclick();
});"
DT::datatable(
data = v$data,
editable = TRUE,
rownames = FALSE,
selection = list(mode = 'none'),
callback = JS(js),
options = list(
searching = FALSE,
paging = FALSE,
ordering = FALSE,
info = FALSE,
autoWidth = TRUE
))
})
#when there is any edit to a cell, write that edit to the initial dataframe
#check to make sure it's positive, if not convert
observeEvent(input$my_datatable_cell_edit, {
#get values
info = input$my_datatable_cell_edit
i = as.numeric(info$row)
j = as.numeric(info$col)
k = as.numeric(info$value)
if(k < 0){ #convert to positive if negative
k <- k * -1
}
#write values to reactive
v$data[i,j] <- k
})
#render plot
output$my_plot <- renderPlot({
req(input$go) #require the input button to be non-0 (ie: don't load the plot when the app first loads)
isolate(v$data) %>% #don't react to any changes in the data
ggplot(aes(x,y))
geom_point()
geom_smooth(method = "lm")
})
}
# Run the application
shinyApp(ui = ui, server = server)
EDIT
I created a js object to click just once and enter the values. Note that after a double click, the cell is approximately the way I would like. I tried to add a setTimeout(), but to no avail. I just wanted it to fill a bigger space and not have scroll. This way:
As if it were Excel/ SPSS or similar:
CodePudding user response:
Firstly, to remove up/down arrows when cell editing and the (blue) inner border that appears when double clicking on the cell, custom css should be added in the ui as follows:
tags$style(HTML("table.dataTable tr td input:focus {outline: none; background: none; border:none;};
table.dataTable tr td input[type=number]::-webkit-inner-spin-button,
input[type=number]::-webkit-outer-spin-button {
-webkit-appearance: none;
margin: 0;
}"))
Then, to remove the numbers you can simply initialise the data frame with NAs instead of zeros:
v <- reactiveValues(data = {
data.frame(x = NA ,y = NA) %>%
add_row(x = rep(NA, 30) ,y = rep(NA, 30))
})
To add a green border around cells when the user is editing the values in them the following JS code should be added:
callback = JS("$('table').on('dblclick', 'td', function() {
$(this).css('border', '1px solid green');
});")
Putting all that together:
library(shiny)
library(shinydashboard)
library(DT)
library(tidyverse)
header <- dashboardHeader(title = "Dashboard", titleWidth = 300)
sidebar <- dashboardSidebar(
width = 300,
sidebarMenu(
menuItem(
text = "Menu 1",
tabName = "menuid1",
icon = icon("chart-line")))
)
body <- dashboardBody(
tabItems(
tabItem(
tabName = "menuid1",
titlePanel(
title = HTML("<text style='background-color:#008cba; color:#f2f2f2;'>Analysis</text>")),
fluidPage(
tags$style(HTML("table.dataTable tr td input:focus {outline: none; background: none; border:none;};
table.dataTable tr td input[type=number]::-webkit-inner-spin-button,
input[type=number]::-webkit-outer-spin-button {
-webkit-appearance: none;
margin: 0;
};")),
column(
id = "meuprimeiromenuid1",
width = 12,
column(
id = "correl",
width = 1,
DT::DTOutput("my_datatable"),
actionButton("go", label = "Plot Data")),
column(
id = "correlplot",
width = 6,
plotOutput("my_plot"))
))
))
)
ui <- dashboardPage(header, sidebar, body)
server <- function(input, output) {
#initialize a blank dataframe
v <- reactiveValues(data = {
data.frame(x = NA ,y = NA) %>%
add_row(x = rep(NA, 30) ,y = rep(NA, 30))
})
callback = JS("$('table').on('dblclick', 'td', function() {
$(this).css('border', '1px solid green');
});")
#output the datatable based on the dataframe (and make it editable)
output$my_datatable <- DT::renderDataTable({
DT::datatable(
data = v$data,
editable = TRUE,
rownames = FALSE,
selection = list(mode = 'none'),
callback = callback,
options = list(
searching = FALSE,
paging = FALSE,
ordering = FALSE,
info = FALSE,
autoWidth = TRUE
))
})
#when there is any edit to a cell, write that edit to the initial dataframe
#check to make sure it's positive, if not convert
observeEvent(input$my_datatable_cell_edit, {
#get values
info = input$my_datatable_cell_edit
i = as.numeric(info$row)
j = as.numeric(info$col) 1
k = as.numeric(info$value)
if (!is.na(k) & k < 0) { #convert to positive if negative
k <- k * -1
}
#write values to reactive
v$data[i,j] <- k
})
#render plot
output$my_plot <- renderPlot({
# browser()
req(input$go) #require the input button to be non-0 (ie: don't load the plot when the app first loads)
isolate(v$data) %>% #don't react to any changes in the data
drop_na() %>%
ggplot(aes(x,y))
geom_point()
geom_smooth(method = "lm")
})
}
# Run the application
shinyApp(ui = ui, server = server)
Also, two additional notes here:
(1) In the cell edit observe event, the variable regarding columns (j) should be increased by 1 given that column count begins at 0 and rownames are disabled.
(2) Given that now the data frame has its values initialised with NA adding appropriate checks for cell editing and when creating/rendering the plot might be required. For example, I've added a command to drop any row that contains value(s) that are NA before creating the plot (drop_na).