I have an adjacency 2D array which has loops meaning the element $a_ii$ is not zero. When trying to plot it with the networkx it messes it up and extra nodes show up! Any suggestions on how to fix this? Here's a small working example.
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
A = np.array([[4, 5, 6],[2, 1, 3],[5, 10, 3]])
G = nx.from_numpy_matrix(A, create_using=nx.DiGraph)
layout = nx.spring_layout(G)
nx.draw_circular(G, with_labels=True)
for edge in G.edges(data="weight"): # edges thickness to be based on the weight
nx.draw_networkx_edges(G, layout, edgelist=[edge], width=edge[2])
plt.show()
CodePudding user response:
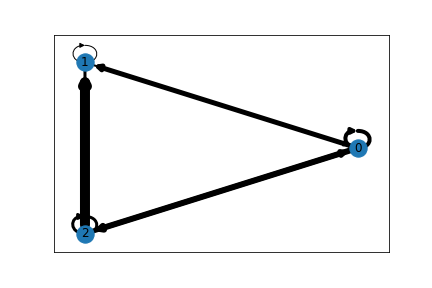
Whenever you have layouts not lining up, check each call to draw is using the same pos parameter. In your for loop, you are using the non-cirucular one. But you are also creating 2 graphs since draw_circular also draws a graph.
Try something like this. Also, you don't necessarily need a for loop to do this.
pos = nx.circular_layout(G)
wt = widths = nx.get_edge_attributes(G, 'weight')
nx.draw_networkx_edges(G, pos, width=list(wt.values()))
nx.draw_networkx_nodes(G, pos)
nx.draw_networkx_labels(G, pos)
Output: