In R, one can easily display eigenvectors in PCA ordination plots using the autoplot function from the ggfortify package, as per the sample code below:
library(ggfortify)
library(ggrepel)
data(iris)
df <- iris[1:4]
pca_res <- prcomp(df, scale. = TRUE)
plt <- plot(autoplot(
pca_res,
data = iris,
colour = 'Species',
label.size = 3,
loadings = TRUE,
loadings.colour = "red",
loadings.label = TRUE,
loadings.label.repel = TRUE,
loadings.label.size = 3
))
plt
With such code, it is possible to control the colours of eigenvectors, however I could not find any way of changing the line width of the eigenvectors.
Could someone tell how to do that?
Thanks in advance!
CodePudding user response:
I don't think you can do this directly within the function call, but you can do it indirectly after the object is created.
Your code creates the following plot:
plt
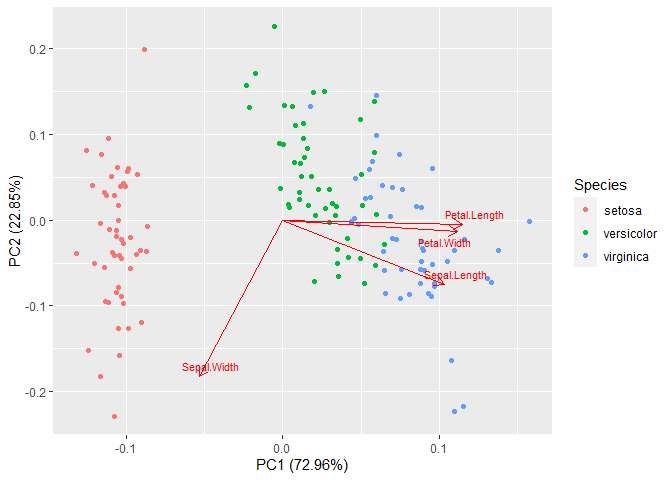
But if we do
plt$layers[[2]]$aes_params$size <- 3
Then we change it to
plt
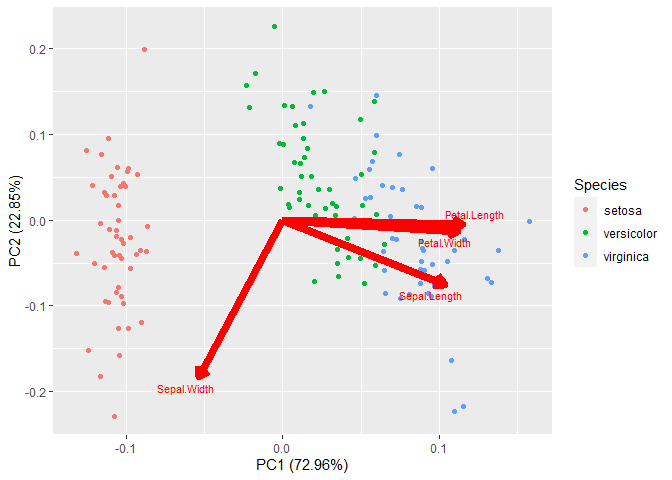
Created on 2022-10-14 with reprex v2.0.2
