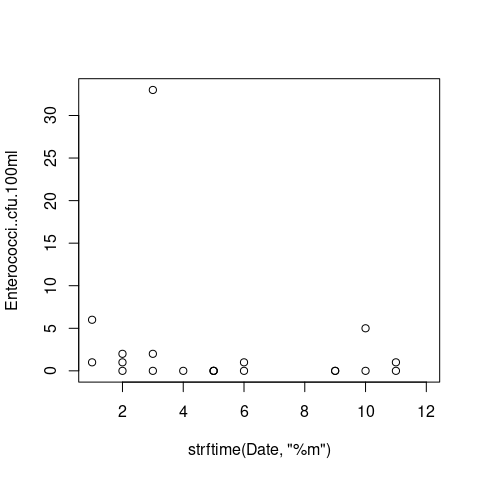
I'm trying to figure out how I can create a scatterplot with the concentration of Enterococci against the Months from this 
Data:
df <- structure(list(Site = c("Avalon Beach", "Avalon Beach", "Avalon Beach",
"Avalon Beach", "Avalon Beach", "Avalon Beach", "Avalon Beach",
"Avalon Beach", "Avalon Beach", "Avalon Beach", "Avalon Beach",
"Avalon Beach", "Avalon Beach", "Avalon Beach", "Avalon Beach",
"Avalon Beach", "Avalon Beach", "Avalon Beach", "Avalon Beach",
"Avalon Beach"), Longitude = c(151.3324, 151.3324, 151.3324,
151.3324, 151.3324, 151.3324, 151.3324, 151.3324, 151.3324, 151.3324,
151.3324, 151.3324, 151.3324, 151.3324, 151.3324, 151.3324, 151.3324,
151.3324, 151.3324, 151.3324), Latitude = c(-33.63587, -33.63587,
-33.63587, -33.63587, -33.63587, -33.63587, -33.63587, -33.63587,
-33.63587, -33.63587, -33.63587, -33.63587, -33.63587, -33.63587,
-33.63587, -33.63587, -33.63587, -33.63587, -33.63587, -33.63587
), Date = c("2018-01-25", "2018-02-07", "2018-02-19", "2018-01-19",
"2018-02-01", "2018-03-08", "2018-03-14", "2018-10-09", "2018-10-31",
"2018-11-07", "2018-11-19", "2018-05-21", "2018-05-25", "2018-06-07",
"2018-06-13", "2018-03-26", "2018-04-10", "2018-05-09", "2018-09-10",
"2018-09-14"), Enterococci..cfu.100ml = c(6L, 0L, 1L, 1L, 2L,
0L, 33L, 5L, 0L, 0L, 1L, 0L, 0L, 1L, 0L, 2L, 0L, 0L, 0L, 0L),
Month = c("January", "February", "February", "January", "February",
"March", "March", "October", "October", "November", "November",
"May", "May", "June", "June", "March", "April", "May", "September",
"September"), Day.of.Week = c("Thursday", "Wednesday", "Monday",
"Friday", "Thursday", "Thursday", "Wednesday", "Tuesday",
"Wednesday", "Wednesday", "Monday", "Monday", "Friday", "Thursday",
"Wednesday", "Monday", "Tuesday", "Wednesday", "Monday",
"Friday")), class = "data.frame", row.names = c(NA, -20L))
CodePudding user response:
The xlim in your error refers to RHS of the formula y ~ x you are using in Enterococci..cfu.100ml. ~ Month, x should be numeric. We can easily strftime the date to get numeric month.
plot(Enterococci..cfu.100ml ~ strftime(Date, '%m'), data=df, xlim=c(1, 12))
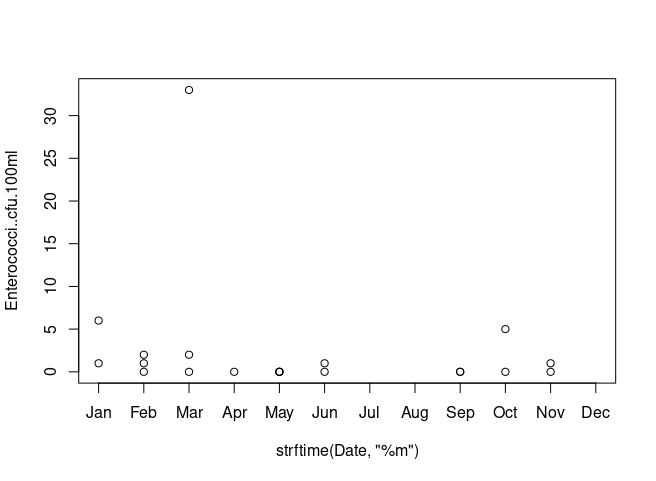
If you want months as x labels, try
plot(Enterococci..cfu.100ml ~ strftime(Date, '%m'), data=df, xlim=c(1, 12), xaxt='n')
axis(1, at=1:12, labels=month.abb[1:12])