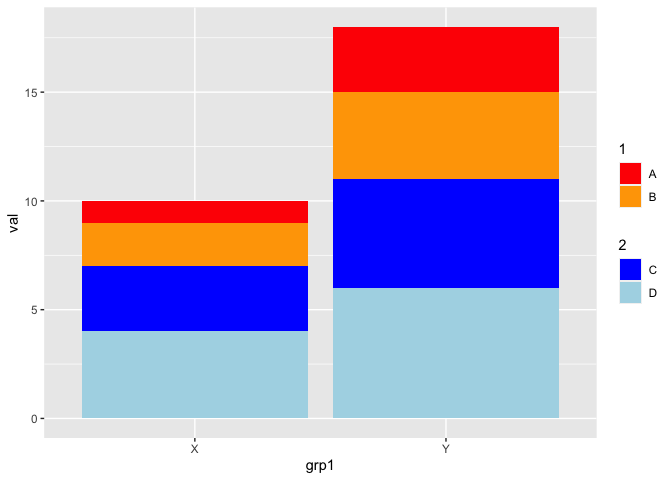
Let say I have below ggplot to draw a stacked bar chart
library(ggplot2)
col_define = c('red', 'orange', 'blue', 'lightblue')
names(col_define) = c('A', 'B', 'C', 'D')
data = rbind(data.frame('grp1' = 'X', 'grp2' = c('A', 'B', 'C', 'D'), 'val' = c(1,2,3,4)), data.frame('grp1' = 'Y', 'grp2' = c('A', 'B', 'C', 'D'), 'val' = c(1,2,3,4) 2))
ggplot(data, aes(x = grp1, fill = grp2, y = val))
geom_bar(stat = 'identity', position = 'stack')
scale_fill_manual(aesthetics = "fill", values = col_define,
breaks = names(col_define))
It is placing all colour in a single legend. However in my case, I basically have 2 groups of colour i.e. one for A & B and second for C & D
I was looking into a similar discussion in ggplot2: Divide Legend into Two Columns, Each with Its Own Title, where there is an approach to group colours of legend using package ggnewscale or relayer
However it looks like, this approach can only be applied in ordinary bar chart, where geom_bar can be called multiple times.
On the contrary, geom_bar, in my case, can't be called multiple times, as it is an whole object
I am looking for some way to use ggnewscale or relayer package in my stack bar chart to group colours in the legend.
As @stefan suggested in one of the answers, a possible way to use geom_col.
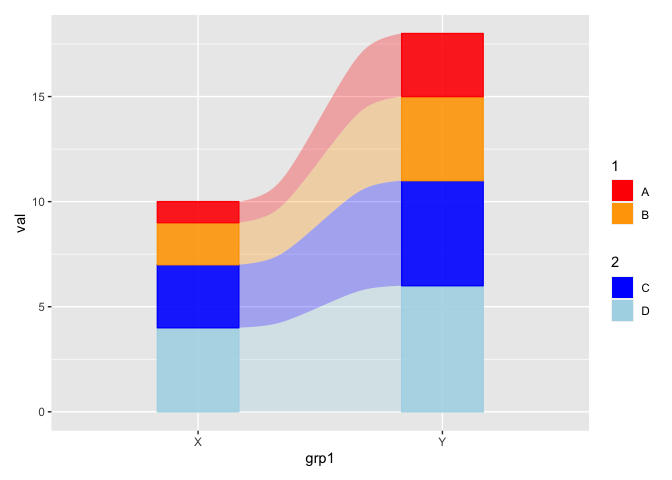
However I found that this approach is fairly restrictive, as I cant apply this method for alluvial plot with the same data as below
library(ggalluvial)
ggplot(data,
aes(x = grp1, stratum = grp2, alluvium = grp2,
y = val,
fill = grp2))
geom_flow(aes(fill = grp2), alpha = .3)
geom_stratum(aes(color = grp2), alpha = .9)
scale_fill_manual(values = col_define, breaks = names(col_define))
Is there a more general approach to group colours in legend?
CodePudding user response:
As the additional information provided asks for a different problem IMHO a second answer is appropriate. Basically it builds on my first answer in that I use ggnewscale to create a grouped legend for a barchart. In second step I extract this legend via cowplot::get_legend to add it to the alluvial plot via patchwork. Once more, this it not elegant but IMHO the easiest way to achieve the desired result:
Note: I tried using ´ggnewscalewithggalluvial` but it seems that the latter is special and a bit stubborn. (; That's why I switched to a different approach.
library(ggplot2)
library(ggnewscale)
library(cowplot)
library(ggalluvial)
library(patchwork)
# Create a grouped legend
p <- ggplot(data, aes(x = grp1, group = grp2, y = val))
geom_col(aes(fill = grp2))
scale_fill_manual(values = col_define, breaks = c("A", "B"), name = "1")
new_scale_fill()
geom_col(aes(fill = grp2))
scale_fill_manual(values = col_define, breaks = c("C", "D"), name = "2")
p_legend <- cowplot::get_legend(p)
# Alluvial plot without legend
p_alluvial <- ggplot(data,
aes(x = grp1, stratum = grp2, alluvium = grp2,
y = val,
fill = grp2))
geom_flow(aes(fill = grp2), alpha = .3)
geom_stratum(aes(color = grp2), alpha = .9)
scale_fill_manual(values = col_define, breaks = names(col_define), aesthetics = c("color", "fill"))
guides(color = "none", fill = "none")
# Alluvial plot with legend via patchwork
p_alluvial p_legend plot_layout(widths = c(10, 1))
#> Warning: `spread_()` was deprecated in tidyr 1.2.0.
#> ℹ Please use `spread()` instead.
#> ℹ The deprecated feature was likely used in the ggalluvial package.
#> Please report the issue at
#> <https://github.com/corybrunson/ggalluvial/issues>.
#> Warning: The `.dots` argument of `group_by()` is deprecated as of dplyr 1.0.0.
#> ℹ The deprecated feature was likely used in the dplyr package.
#> Please report the issue at <https://github.com/tidyverse/dplyr/issues>.
CodePudding user response:
Not elegant but of course could you simply add a second geom_col on top of the first to get two legends:
library(ggplot2)
library(ggnewscale)
ggplot(data, aes(x = grp1, group = grp2, y = val))
geom_col(aes(fill = grp2))
scale_fill_manual(values = col_define, breaks = c("A", "B"), name = "1")
new_scale_fill()
geom_col(aes(fill = grp2))
scale_fill_manual(values = col_define, breaks = c("C", "D"), name = "2")