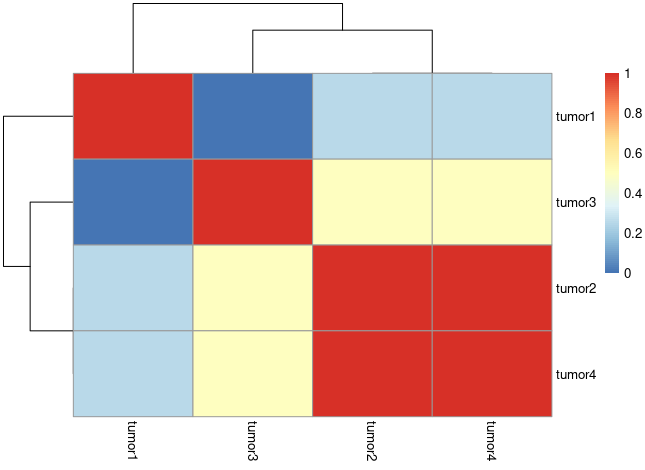
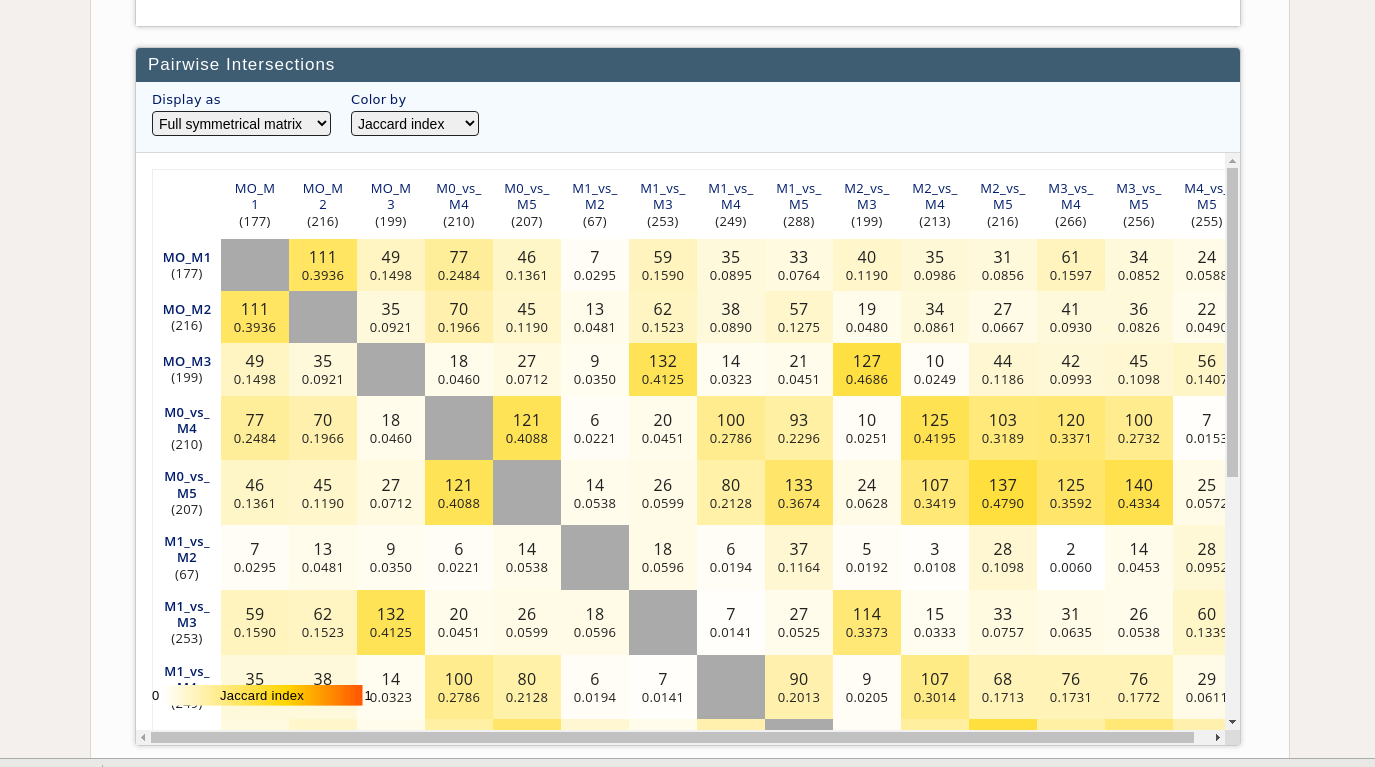
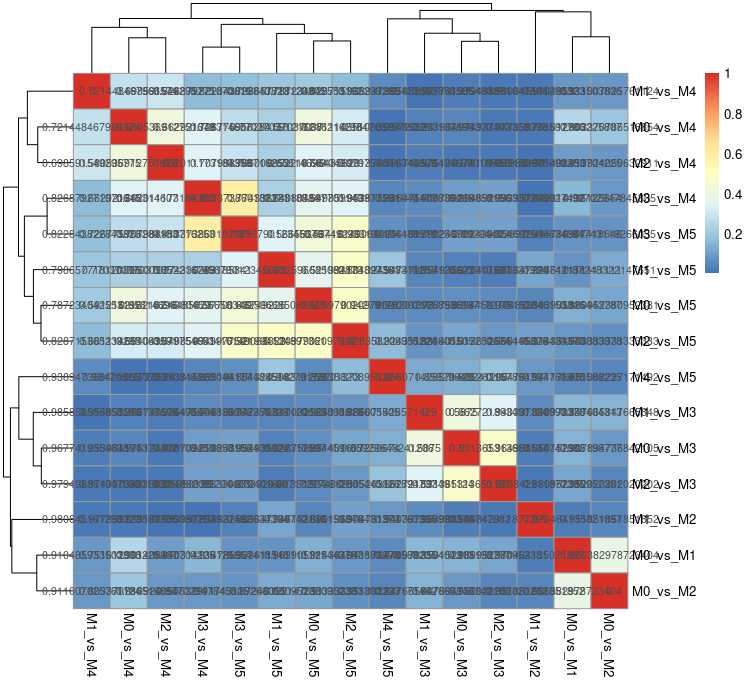
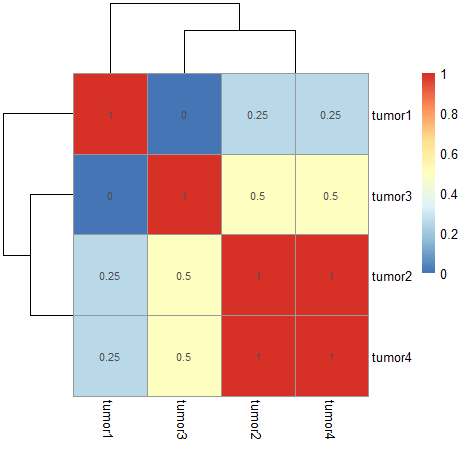
Im trying to calculate and plotting the jaccard index the same
My dummy data and code
df <- data.frame(tumor_type = c("tumor1", "tumor1", "tumor1", "tumor2", "tumor2", "tumor3", "tumor4", "tumor4"), genes = c("geneA", "geneB", "geneC", "geneA", "geneD", "geneD", "geneA", "geneD"))
D1 <- vegdist(unclass(table(df$tumor_type,df$genes)), method = "jaccard")
pheatmap(1-as.matrix(D1))
For my actual data I tried an online tool i get this
My objective is to get similar such as showing the element present in each sub-type and their jacard score
Small subset of my data
structure(list(Comparison = c("M0_vs_M1", "M0_vs_M1", "M0_vs_M1",
"M0_vs_M1", "M0_vs_M1", "M0_vs_M1", "M4_vs_M5", "M4_vs_M5", "M4_vs_M5",
"M4_vs_M5", "M4_vs_M5", "M4_vs_M5"), Term = c("GO:0042742", "GO:0009617",
"GO:0006959", "GO:0006956", "GO:0006958", "GO:0002455", "GO:0048863",
"GO:0048864", "GO:0001667", "GO:0048483", "GO:0051129", "GO:0010977"
)), row.names = c(1L, 2L, 3L, 4L, 5L, 6L, 3266L, 3267L, 3268L,
3269L, 3270L, 3271L), class = "data.frame")
My output
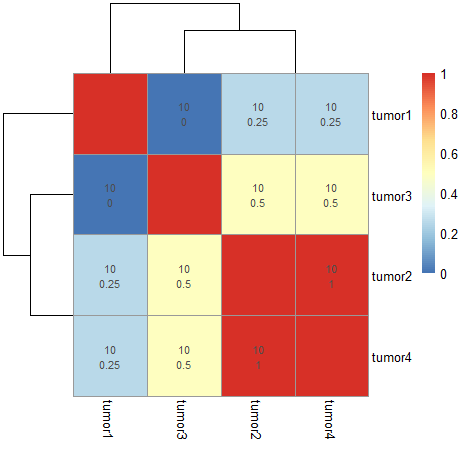
CodePudding user response:
You may add values by using display_numbers = ... argument inside pheatmap().
pheatmap(1-as.matrix(D1), display_numbers = 1-as.matrix(D1))
D2 <- matrix(ncol = 4, nrow = 4)
D2[lower.tri(D2)] <- 1 - D1
D2 <- matrix(D2 % % t(D2), ncol = 4)
diag(D2) <- NA
D2
D3 <- matrix(rep(10, 16), ncol = 4)
diag(D3) <- NA
D3
D4 <- matrix(paste(D3,D2, sep ="\n"), ncol = 4)
diag(D4) <- ""
pheatmap(1-as.matrix(D1), display_numbers = D4)