So I have a bunch of studies and I want to plot the regression lines obtained from all of them.
I have a set of slope and intercept values, and x limits for each line, but I need to set the x limits for each line separately as they're all different...
I've racked my brain for days but can't figure out how I can achieve this using ggplot, if I can give a list of xlim values to pass to the slope and intercept values?
Here's a subset of my dataframe...
empirical_studies <- tibble(slope = c(-1.52, -1.42, -1.56, -1.57, -1.57, -1.68, -1.67, -1.6, -1.73, -1.69, -1.79), intercept = c(12.07, 11.29, 12.21, 12.26, 12.14, 12.5, 12.58, 12.28, 12.72, 12.53, 12.29), xmin = c(9.00, 6.00, 6.00, 6.00, 6.00, 9.00, 7.00, 7.00, 7.00, 7.00, 18.00), xmax = c(26.00 59.00, 53.00, 53.00, 53.00, 63.00, 67.00, 64.00, 64.00, 64.00, 50.00))
Would really appreciate any help!
I didn't really try much to be honest, I know I can't pass in a set of xlim values... I tried writing a loop (below) but this doesn't seem to work either:
plot = ggplot(empirical_studies, aes(x=empirical_studies$diameter_stem_average, y=empirical_studies$density))
for (i in 1:(length(empirical_studies)-1)){
plot=plot geom_abline(slope = empirical_studies$slope[i], intercept = empirical_studies$intercept[i])
xlim(empirical_studies$min_dq[i], empirical_studies$max_dq[i])
i <- i 1
}
print(plot)
CodePudding user response:
If you want to plot separate line segments you can use geom_line() with a start and end point.
Since you already have the x values of the min and max, you can simply use the slope and intercept to calculate the y value for those points and then plot it. For these manipulations you need to uniquely identify each curve so I just applied a number for each, but you could use more informative naming depending on your data.
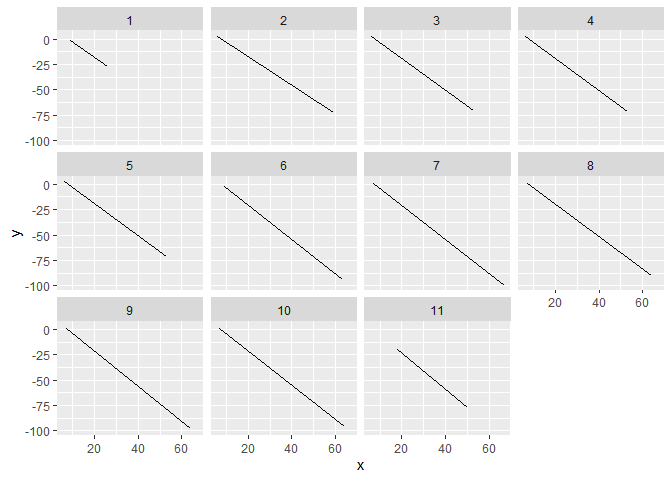
I show two options below depending on if you want to overplot everything or if you want separate plots for each. For the faceted option, you can specify sacles = "free" to let each facet rescale the axes individually. Otherwise it picks the widest range that captures all the data.
library(tidyverse)
empirical_studies <- tibble(slope = c(-1.52, -1.42, -1.56, -1.57, -1.57, -1.68, -1.67, -1.6, -1.73, -1.69, -1.79),
intercept = c(12.07, 11.29, 12.21, 12.26, 12.14, 12.5, 12.58, 12.28, 12.72, 12.53, 12.29),
xmin = c(9.00, 6.00, 6.00, 6.00, 6.00, 9.00, 7.00, 7.00, 7.00, 7.00, 18.00),
xmax = c(26.00, 59.00, 53.00, 53.00, 53.00, 63.00, 67.00, 64.00, 64.00, 64.00, 50.00))
# generate list of start and end points for each line
d <- empirical_studies %>%
mutate(study_id = row_number()) %>%
mutate(across(starts_with("x"), ~ slope * . intercept, .names = "y{str_sub(.col, 2, 4)}")) %>%
pivot_longer(starts_with(c("x", "y"))) %>%
separate(name, into = c("var", "position"), sep = 1) %>%
pivot_wider(names_from = var)
d
#> # A tibble: 22 × 6
#> slope intercept study_id position x y
#> <dbl> <dbl> <int> <chr> <dbl> <dbl>
#> 1 -1.52 12.1 1 min 9 -1.61
#> 2 -1.52 12.1 1 max 26 -27.4
#> 3 -1.42 11.3 2 min 6 2.77
#> 4 -1.42 11.3 2 max 59 -72.5
#> 5 -1.56 12.2 3 min 6 2.85
#> 6 -1.56 12.2 3 max 53 -70.5
#> 7 -1.57 12.3 4 min 6 2.84
#> 8 -1.57 12.3 4 max 53 -71.0
#> 9 -1.57 12.1 5 min 6 2.72
#> 10 -1.57 12.1 5 max 53 -71.1
#> # … with 12 more rows
# plotted together
d %>%
ggplot(aes(x, y))
geom_line(aes(color = factor(study_id)))
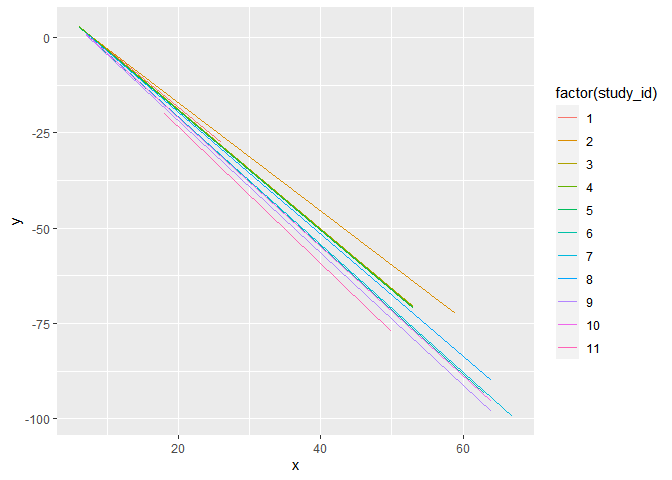
# plotted separately
d %>%
ggplot(aes(x, y))
geom_line()
facet_wrap(~study_id) # can add scales = "free" to scale axes separately
Created on 2022-11-09 with reprex v2.0.2
