I like to make a somewhat easy calculation on the rows of my data frame and used to use .iterrows() but the the operation is very slow. Now I wonder if I can use .apply() to achieve the same thing to get it done faster. It could also be that there is a totally differnt option, which I'm just not aware of or have not thought about.
Here is what I want to do: Assuming the following dataframe
| ID_1 | ID_2 | ... | ID_n | mean | |
|---|---|---|---|---|---|
| 0 | 10 | 15 | ... | 12 | 7 |
| 1 | 20 | 10 | ... | 17 | 21 |
I like to check for each row which element is larger than the mean of the entire row (already given in the mean column). If the value is larger I like to get the part of the ID after the _ (column name) for this entry and finally sum up all the values that are larger than the row mean and safe it to a new column.
Thanks for any help.
I already tried to use
df.apply(lamda row: my_func(row), axis=1)
def my_func(x):
id = str(x.index)
if x[x.name] > (df['mean'].iloc[x.name]):
sum( x )
CodePudding user response:
This works:
d = np.array([ [10,15,12,7],
[20,10,17,21]])
df = pd.DataFrame(d, columns=["ID_1","ID_2","ID_3","mean"])
N = 3
def my_func(row):
s = 0
for i in range(1,N 1):
if row[f"ID_{i}"] > row["mean"]:
s = row[f"ID_{i}"]
return s
df["sum_lrgr_mean"] = df.apply(lambda row: my_func(row), axis=1)
df
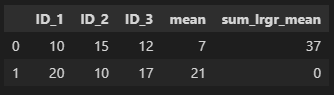
This will produce:
CodePudding user response:
Also, if you want speed, you can move from pandas to numpy arrays as such:
N = np.array(
[
[10, 15, 12],
[20, 10, 17]
]
)
M = np.array(
[
[7],
[21]
]
)
np.sum(N*(N>M),axis=1)
Which will produce this array:
array([37, 0])
CodePudding user response:
Lets use .melt with .loc, .groupby and .join to get your values.
#we need the index to rejoin later
df1 = pd.melt(df,id_vars='mean',ignore_index=False).reset_index()
con = df1['value'].gt(df1['mean']) # your conditional.
df_new = df.join(df1.loc[con].assign(_id=df1['variable'].str.split('_').str[1]
).groupby('index')\
.agg(_id=('_id',list),computed_mean=('value','sum'))
)
print(df_new)
ID_1 ID_2 ID_n mean _id computed_mean
0 10 15 12 7 [1, 2, n] 37.0
1 20 10 17 21 NaN NaN
if we look into df1 & con we can see the records we are interested in.
index mean variable value
0 0 7 ID_1 10
1 1 21 ID_1 20
2 0 7 ID_2 15
3 1 21 ID_2 10
4 0 7 ID_n 12
5 1 21 ID_n 17
print(con)
0 True
1 False
2 True
3 False
4 True
5 False
dtype: bool
Option 2
If you don't need the IDs as a list then a simple sum and mask will do.
df['computed_mean'] = df.mask(df.lt(df['mean'],axis=0)).drop('mean',axis=1).sum(axis=1)
ID_1 ID_2 ID_n mean computed_mean
0 10 15 12 7 37.0
1 20 10 17 21 0.0