I'm new with Python. I have to create a new rv U(t). Assuming that Z has a standard normal distribution c = 1.57, I have that:
U(t) = 0 if Z(t) <= c
U(t) = Φ(Z(t)) Z(t) > c
Where Φ(·) is the cdf of the standard normal distribution N(0, 1).
I start sampling random numbers from the normal distribution and I create an array of zeros:
z = np.random.normal(0, 1, 100)
u = np.zeros([1, 100])
Then, I write the following loop:
for i in list(range(1, 101, 1)):
if z[:i] < c:
u[:i] = 0
else z[:i] > c:
u[:i] = norm.cdf(z[:i], loc=0, scale=1)
However, there is something wrong. I got this error:
File "<ipython-input-837-1cbdd0641a75>", line 4
else z[:i] > c:
^
SyntaxError: invalid syntax
Can someone please help me find out where the error is? Or suggest another way to deal with the problem?
Thank you!
CodePudding user response:
Use the power of numpy!
z = np.random.normal(0, 1, 100)
u = np.zeros(z.shape)
Since you initialized u to zeros, you don't need to do anything for the z <= c cases. For the others, you can use numpy's logical indexing to only set the elements that fulfill the condition
# Get only the elements of z where z > c
z_filt = z[z > c]
# Calculate the norm.cdf at these values of z,
norm_vals = norm.cdf(z_filt, loc=0, scale=1)
# Assign those to the elements of u where z > c
u[z > c] = norm_vals
Of course, you can condense these three lines down to a single line:
u[z > c] = norm.cdf(z[z > c], loc=0, scale=1)
This approach will be significantly faster than iterating over the arrays and setting individual elements.
If you're curious why your code didn't work and how to fix it,
- You don't need to list out a
rangeto iterate on it. Justfor i in range(100)is good enough z[:i] < cwill give an array containingiboolean values. Putting anifcondition on that will give you an error:ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all(). I suspect you meant to doz[i] < cu[:i] = 0will set all elements ofufrom the start to theith index to zero. You probably only wantedu[i] = 0. This is really not even necessary since you already initializeduto be zeroselse <condition>doesn't work. You wantelif <condition>. Although you don't really need that here, since you could just doif z[i] > cand that's the only condition you need.
for i in range(100):
if z[i] > c:
u[i] = norm.cdf(z[i], loc=0, scale=1)
Comparing the runtimes of these two approaches:
import timeit
import numpy as np
from scipy.stats import norm
from matplotlib import pyplot as plt
def f_numpy(size):
z = np.random.normal(0, 1, size)
u = np.zeros(z.shape)
u[z > c] = norm.cdf(z[z > c], loc=0, scale=1)
return u
def f_loopy(size):
z = np.random.normal(0, 1, size)
u = np.zeros(z.shape)
for i in range(size):
if z[i] > c:
u[i] = norm.cdf(z[i], loc=0, scale=1)
return u
c = 0
sizes = [10, 100, 1000, 10_000, 100_000]
times = np.zeros((len(sizes), 2))
for i, s in enumerate(sizes):
times[i, 0] = timeit.timeit('f_numpy(s)', globals=globals(), number=100) / 100
print(">")
times[i, 1] = timeit.timeit('f_loopy(s)', globals=globals(), number=10) / 10
print(".")
fig, ax = plt.subplots()
ax.plot(sizes, times[:, 0], label="numpy")
ax.plot(sizes, times[:, 1], label="loopy")
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_xlabel('Array size')
ax.set_ylabel('Time per function call (s)')
ax.legend()
fig.tight_layout()
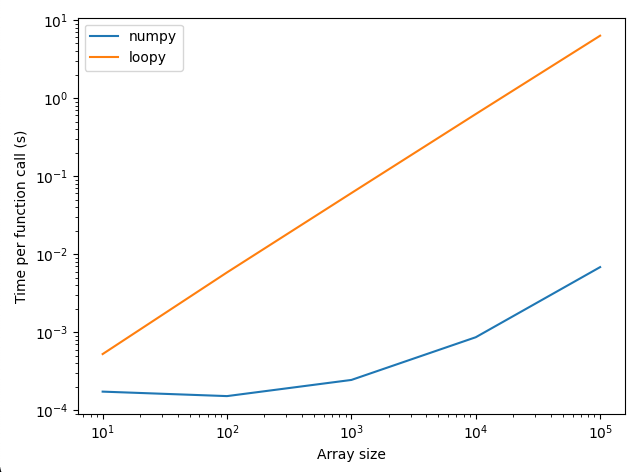
we get the following plot, which shows that the numpy approach is orders of magnitude faster than the loopy approach.