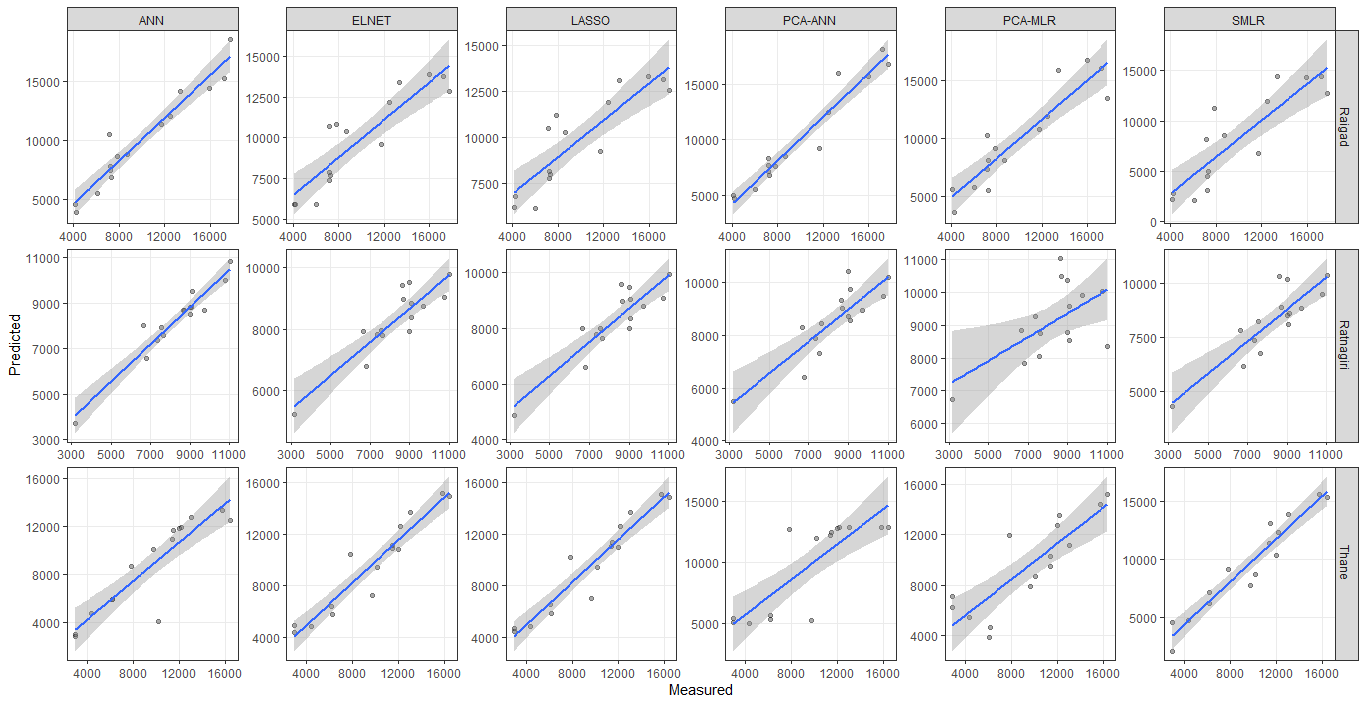
I am using your facet_grid2 function from ggh4x to make both x and y-axis scales to be free like
ggplot(data_calibration, aes(Observed,Predicted))
geom_point(color="black",alpha = 1/3)
facet_grid2(Station ~ Method, scales="free", independent = "all")
xlab("Measured")
ylab("Predicted")
theme_bw()
geom_smooth(method="lm")
theme(panel.grid.minor = element_blank())
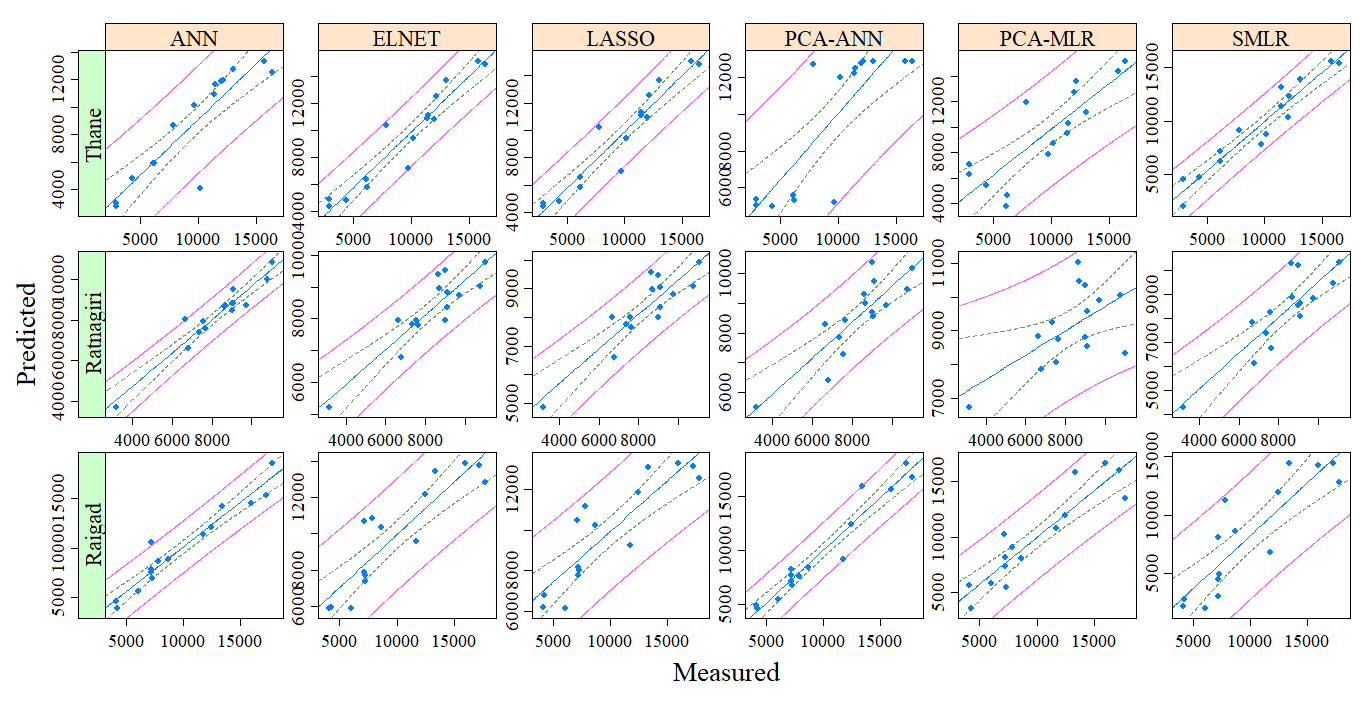
Now how can I add the 95% confidence interval of prediction to this plot like the following plot
Data
data_calibration = structure(list(Observed = c(17229L, 15964L, 13373L, 17749L, 12457L,
7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L, 7197L, 7220L,
7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L, 11465L,
11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L, 10759L,
9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L, 3183L,
9078L, 8688L, 11023L, 9000L, 9001L, 17229L, 15964L, 13373L, 17749L,
12457L, 7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L, 7197L,
7220L, 7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L,
11465L, 11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L,
10759L, 9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L,
3183L, 9078L, 8688L, 11023L, 9000L, 9001L, 17229L, 15964L, 13373L,
17749L, 12457L, 7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L,
7197L, 7220L, 7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L,
11465L, 11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L,
10759L, 9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L,
3183L, 9078L, 8688L, 11023L, 9000L, 9001L, 17229L, 15964L, 13373L,
17749L, 12457L, 7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L,
7197L, 7220L, 7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L,
11465L, 11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L,
10759L, 9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L,
3183L, 9078L, 8688L, 11023L, 9000L, 9001L, 17229L, 15964L, 13373L,
17749L, 12457L, 7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L,
7197L, 7220L, 7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L,
11465L, 11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L,
10759L, 9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L,
3183L, 9078L, 8688L, 11023L, 9000L, 9001L, 17229L, 15964L, 13373L,
17749L, 12457L, 7166L, 7842L, 8675L, 11718L, 6049L, 4232L, 4126L,
7197L, 7220L, 7284L, 16410L, 15772L, 12166L, 11997L, 7827L, 13034L,
11465L, 11409L, 10165L, 9702L, 2942L, 2940L, 4361L, 6197L, 6144L,
10759L, 9720L, 8631L, 7354L, 7640L, 6653L, 7551L, 6791L, 9093L,
3183L, 9078L, 8688L, 11023L, 9000L, 9001L), Station = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("Raigad",
"Ratnagiri", "Thane "), class = "factor"), Method = structure(c(6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L), .Label = c("ANN",
"ELNET", "LASSO", "PCA-ANN", "PCA-MLR", "SMLR"), class = "factor"),
Predicted = c(14463L, 14285L, 14452L, 12765L, 11917L, 8143L,
11251L, 8611L, 6789L, 2059L, 2787L, 2201L, 3062L, 4508L,
4975L, 15357L, 15605L, 12326L, 10377L, 9113L, 13926L, 13142L,
11407L, 8711L, 7801L, 2064L, 4563L, 4725L, 6247L, 7170L,
9492L, 8857L, 10323L, 7389L, 6776L, 7842L, 8261L, 6156L,
8627L, 4326L, 8094L, 8897L, 10370L, 10214L, 8548L, 16043L,
16671L, 15831L, 13463L, 11921L, 10239L, 9110L, 8090L, 10794L,
5826L, 3621L, 5639L, 7364L, 8152L, 5515L, 15182L, 14370L,
13559L, 12748L, 11936L, 11125L, 10313L, 9502L, 8691L, 7879L,
7068L, 6257L, 5445L, 4634L, 3822L, 10045L, 9911L, 11038L,
9255L, 8736L, 8848L, 8063L, 7847L, 8538L, 6744L, 9583L, 10474L,
8343L, 10353L, 8791L, 13185L, 13331L, 13099L, 12557L, 11898L,
10474L, 11199L, 10255L, 9251L, 6148L, 6795L, 6166L, 7775L,
8157L, 7990L, 14843L, 15086L, 12585L, 10987L, 10193L, 13663L,
11317L, 11071L, 9392L, 6991L, 4484L, 4667L, 4846L, 5830L,
6577L, 9085L, 8802L, 9570L, 7770L, 7652L, 8006L, 7995L, 6599L,
9050L, 4876L, 8360L, 8981L, 9931L, 9479L, 8009L, 13775L,
13890L, 13416L, 12851L, 12141L, 10693L, 10834L, 10372L, 9585L,
5914L, 5930L, 5922L, 7854L, 7407L, 7697L, 14941L, 15174L,
12572L, 10817L, 10412L, 13705L, 11154L, 10886L, 9448L, 7215L,
4389L, 4875L, 4809L, 5747L, 6385L, 9034L, 8749L, 9410L, 7820L,
7798L, 7940L, 7957L, 6803L, 8844L, 5227L, 8369L, 8972L, 9789L,
9514L, 7940L, 15309L, 14477L, 14219L, 18581L, 12084L, 10550L,
8666L, 8812L, 11415L, 5566L, 3928L, 4592L, 7861L, 7489L,
6903L, 12509L, 13366L, 11956L, 11880L, 8711L, 12768L, 11690L,
10922L, 4101L, 10106L, 2811L, 2979L, 4785L, 5944L, 5901L,
10007L, 8710L, 8688L, 7383L, 7575L, 8047L, 7938L, 6585L,
9517L, 3729L, 8816L, 8704L, 10847L, 8812L, 8493L, 18115L,
15670L, 15931L, 16804L, 12450L, 7701L, 7588L, 8450L, 9205L,
5477L, 4666L, 4948L, 8262L, 7095L, 6798L, 12902L, 12883L,
12864L, 12788L, 12690L, 12896L, 12491L, 12199L, 11982L, 5213L,
5357L, 5053L, 5013L, 5321L, 5596L, 9467L, 8931L, 9305L, 7867L,
8427L, 8282L, 7291L, 6396L, 9725L, 5509L, 8545L, 8997L, 10171L,
10389L, 8700L)), class = "data.frame", row.names = c(NA,
-270L))
CodePudding user response:
In short, the geom_smooth function only calculates confidence intervals. To get prediction intervals as well, these should be calculated outside of ggplot and passed in. This is a bit of a long way of coding, but hopefully you can see that predict is called twice on the lm model, once to produce two columns of confidence intervals, once to produce two columns of prediction intervals. These are passed on to geom_ribbons:
library(ggh4x)
library(tidyverse)
data_calibration |>
group_by(Station, Method) |>
nest() |>
mutate(model = map(data, ~ lm(Predicted ~ Observed, data = .x))) |>
mutate(fit = map2(model, data, ~ as.tibble(
predict(.x, interval = "conf"), new_data = tibble(Observed = seq(min(
data$Observed, max(data$Observed), 100
)))
)),
pred = map2(model, data, ~ as.tibble(
predict(.x, interval = "pred", new_data = tibble(Observed = seq(
min(data$Observed, max(data$Observed), 100)
)))
))) |>
unnest(c(data, fit, pred), names_sep = "_") |>
ggplot(aes(data_Observed, data_Predicted))
geom_point(color = "black", alpha = 1 / 3)
facet_grid2(Station ~ Method, scales = "free", independent = "all")
xlab("Measured")
ylab("Predicted")
theme_bw()
geom_smooth(method = "lm", se = FALSE)
geom_ribbon(aes(ymax = fit_upr, ymin = fit_lwr),
colour = "green",
fill = NA)
geom_ribbon(aes(ymax = pred_upr, ymin = pred_lwr),
colour = "red",
fill = NA)
theme(panel.grid.minor = element_blank())
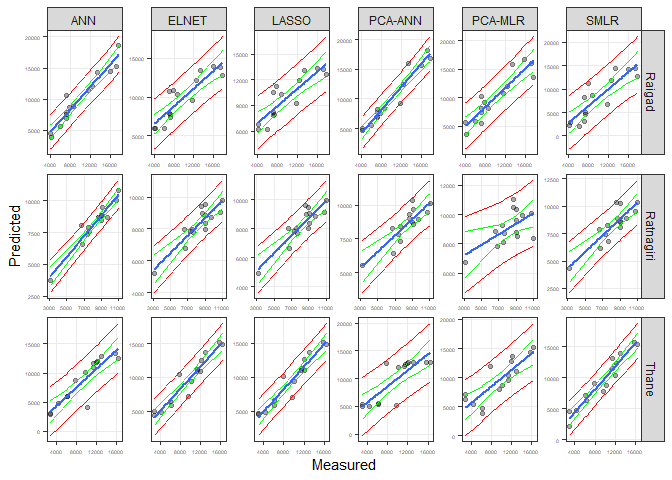
I would welcome a tidier answer! One would be to create a new stat_predict layer function, which is a little tricky but not impossible.
Edit - that thing I said was perhaps a good idea, maybe it is!
Out of curiosity, I thought worth making a stat_predict function. Source the code from this gist and then the simple code will work with above data:
# To source new function, either...
source("https://gist.githubusercontent.com/andrewbaxter439/b508a60786f8af3c0be7b381a667ae07/raw/f7f4672222f0b1024cf6bf536ed7f6059867b4f2/stat_predict.R")
# or devtools::source_gist("b508a60786f8af3c0be7b381a667ae07")
ggplot(data_calibration, aes(Observed,Predicted))
geom_point(color="black",alpha = 1/3)
facet_grid2(Station ~ Method, scales="free", independent = "all")
xlab("Measured")
ylab("Predicted")
theme_bw()
geom_smooth(method="lm", se = FALSE)
stat_smooth(method = "lm", geom = "ribbon", fill = NA, colour = "green")
stat_predict(method = "lm", geom = "ribbon", fill = NA, colour = "red")
theme(panel.grid.minor = element_blank())