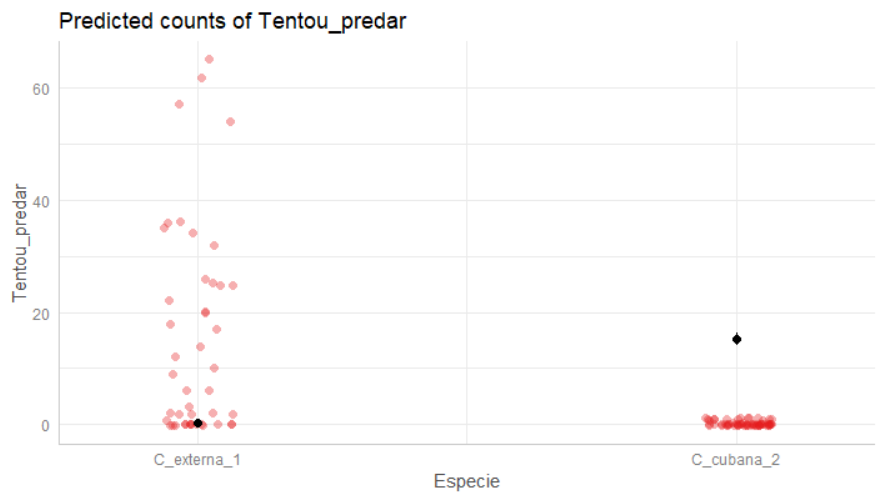
I'd been trying to plot a glm model with ggeffects::ggpredict() but I'm getting an error. I've tried to change the data type of the variable "Especie" to factor, but it didn't work.
library(dplyr)
library(readr)
library(ggplot2)
library(ggeffects)
# Read my data
ds <- structure(list(Especie = c("C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1", "C_externa_1",
"C_externa_1", "C_externa_1", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2", "C_cubana_2",
"C_cubana_2", "C_cubana_2", "C_cubana_2"),
Tentou_predar = c(3L, 25L, 20L, 36L, 12L, 0L, 1L, 10L,
0L, 14L, 2L, 0L, 0L, 0L, 0L, 32L, 0L, 0L, 25L, 0L, 2L, 2L, 35L,
0L, 0L, 0L, 22L, 0L, 2L, 9L, 54L, 57L, 26L, 17L, 18L, 34L, 2L,
0L, 20L, 25L, 6L, 65L, 36L, 6L, 62L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 1L, 0L, 0L, 1L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 1L, 0L, 1L, 1L, 0L, 1L, 0L, 0L,
1L, 0L, 0L, 1L, 0L, 0L, 1L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 1L)), class = "data.frame", row.names = c(NA,
-105L))
str(ds)
# Create a glm model and plot it
m_Pred <- glm(Tentou_predar ~ Especie, data = ds,
family = "poisson")
df_gg <- ggeffects::ggpredict(m_Pred, terms = "Especie [all]")
df_gg$x_1 <- 1 (readr::parse_number(as.character(df_gg$group)) - 2) * 0.05
df_gg %>% plot(add.data = TRUE)
The mean point is wrong, the high is values in "C_cubana_2" and not in "C_externa_1"!
# But the mean and bars are wrong:
df_gg %>% group_by(x) %>%summarize(predicted=mean(predicted))
# # A tibble: 2 x 2
# x predicted
# <fct> <dbl>
# 1 C_cubana_2 0.233
# 2 C_externa_1 15.1
Please any help with it?
CodePudding user response:
You should convert your Especie variable to a factor like this:
library(dplyr)
library(readr)
library(ggplot2)
library(ggeffects)
-105L))
ds$Especie <- as.factor(ds$Especie)
str(ds)
#> 'data.frame': 105 obs. of 2 variables:
#> $ Especie : Factor w/ 2 levels "C_cubana_2","C_externa_1": 2 2 2 2 2 2 2 2 2 2 ...
#> $ Tentou_predar: int 3 25 20 36 12 0 1 10 0 14 ...
# Create a glm model and plot it
m_Pred <- glm(Tentou_predar ~ Especie, data = ds,
family = "poisson")
df_gg <- ggeffects::ggpredict(m_Pred, terms = "Especie [all]")
df_gg$x_1 <- 1 (readr::parse_number(as.character(df_gg$group)) - 2) * 0.05
df_gg %>% plot(add.data = TRUE)
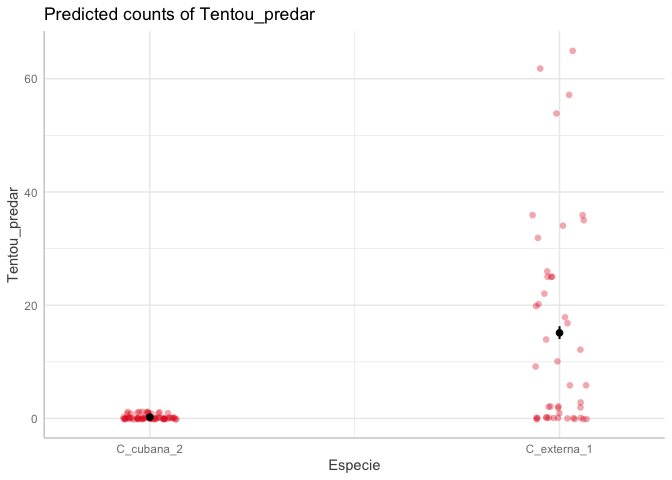
df_gg %>% group_by(x) %>%summarize(predicted=mean(predicted))
#> # A tibble: 2 × 2
#> x predicted
#> <fct> <dbl>
#> 1 C_cubana_2 0.233
#> 2 C_externa_1 15.1
Created on 2022-12-11 with reprex v2.0.2