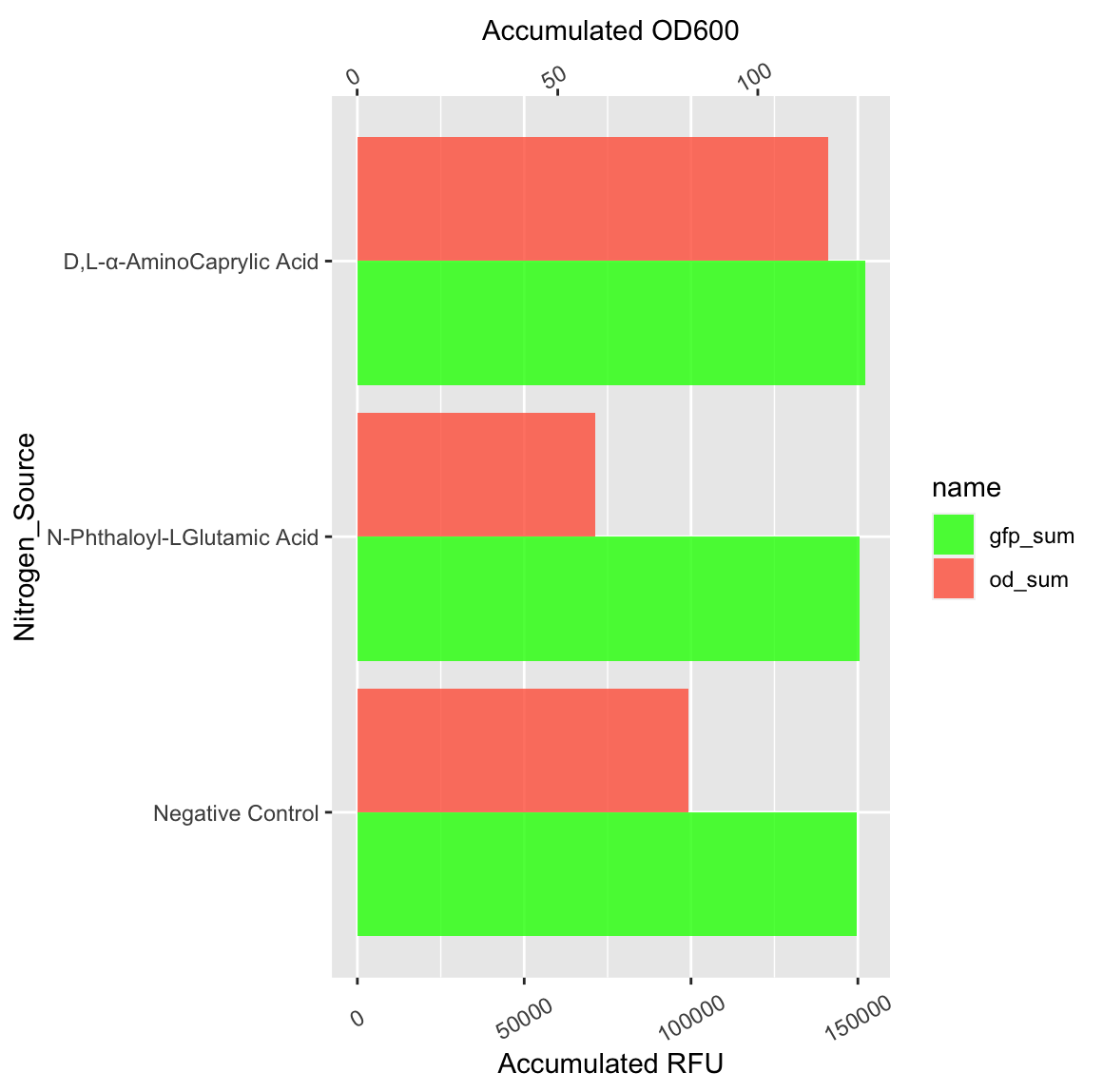
im currently trying to plot some bar charts where there are two values for each groups. As the values are far from each other i've made two axis.
However, i would like the "Accumulated RFU" y-axis to be on top instead of "Accumulated OD600", but so far it's on the bottom.
My script looks like this for the plot:
ggplot(mean_sum_long, aes(x = Nitrogen_Source, y = value, fill = name))
geom_col(position="dodge",alpha=0.8)
scale_y_continuous(
name = "Accumulated RFU",
sec.axis = sec_axis(~./coeff, name="Accumulated OD600"))
scale_fill_manual(values = c("green", "tomato"))
theme(axis.text.x = element_text(angle = 30, vjust = 0.5))
coord_flip()
coeff being 5000.
Data:
structure(list(Nitrogen_Source = structure(c(1L, 1L, 2L, 2L,
3L, 3L), levels = c("Negative Control", "N-Phthaloyl-LGlutamic Acid",
"D,L-α-AminoCaprylic Acid", "α-Amino-NValeric Acid", "L-Tryptophan",
"Methylamine", "L-Cysteine", "Gly-Met", "Ethylenediamine", "Ala-His",
"L-Tyrosine", "Ala-Leu", "Formamide", "D-Lysine", "D-Galactosamine",
"Nitrate", "L-Histidine", "N-Acetyl-DMannosamine", "Ethylamine",
"D-Mannosamine", "Histamine", "N-Acetyl-DGalactosamine", "Biuret",
"D,L-Lactamide", "N-Acetyl-LGlutamic Acid", "Parabanic Acid",
"Met-Ala", "L-Pyroglutamic Acid", "ß-Phenylethylamine", "Glucuronamide",
"D-Glucosamine", "L-Glutamic Acid", "L-Glutamine", "Thymidine",
"L-Isoleucine", "Xanthine", "Nitrite", "Ala-Asp", "Uridine",
"ε-Amino-NCaproic Acid", "L-Citrulline", "D-Alanine", "Thymine",
"L-Phenylalanine", "Ala-Glu", "Uric Acid", "γ-Amino-NButyric Acid",
"Ammonia", "L-Aspartic Acid", "D-Aspartic Acid", "δ-Amino-NValeric Acid",
"Ethanolamine", "L-Threonine", "Putrescine", "L-Methionine",
"Allantoin", "L-Ornithine", "Ala-Thr", "Ala-Gln", "Cytosine",
"Ala-Gly", "Agmatine", "L-Asparagine", "Inosine", "Guanosine",
"Glycine", "N-Acetyl-DGlucosamine", "Adenosine", "Xanthosine",
"L-Leucine", "Uracil", "L-Serine", "L-Homoserine", "Gly-Gln",
"L-Proline", "L-Alanine", "Gly-Asn", "Urea", "Cytidine", "D-Glutamic Acid",
"L-Arginine", "Gly-Glu", "Adenine"), class = "factor"), name = c("gfp_mean",
"od_mean", "gfp_mean", "od_mean", "gfp_mean", "od_mean"), value = c(74851.5,
41.29, 75282.5, 29.6650000000001, 76050, 58.755)), row.names = c(NA,
-6L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
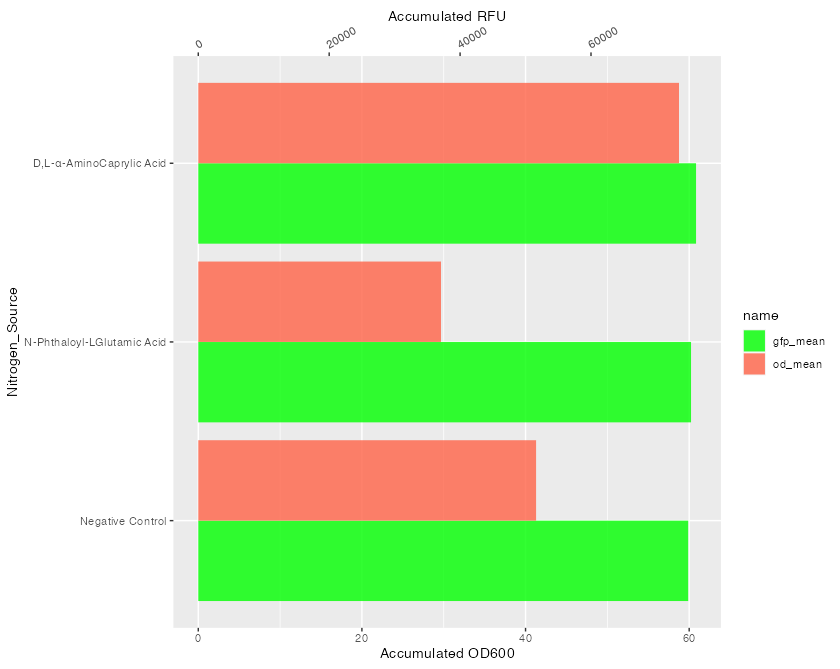
To achieve your desired result you have to use the reverse transformation, transform your gfp_mean values and of course switch the transformation for the secondary axis as well as the names.
Note: At least for the provided example data a coeff of 1250 seems more appropriate.
library(ggplot2)
coeff <- 1250
ggplot(mean_sum_long, aes(x = Nitrogen_Source, y = ifelse(name == "gfp_mean", value / coeff, value), fill = name))
geom_col(position = "dodge", alpha = 0.8)
scale_y_continuous(
name = "Accumulated OD600",
sec.axis = sec_axis(~ . * coeff, name = "Accumulated RFU")
)
scale_fill_manual(values = c("green", "tomato"))
theme(axis.text.x.top = element_text(angle = 30, vjust = 0, hjust = 0))
coord_flip()
CodePudding user response:
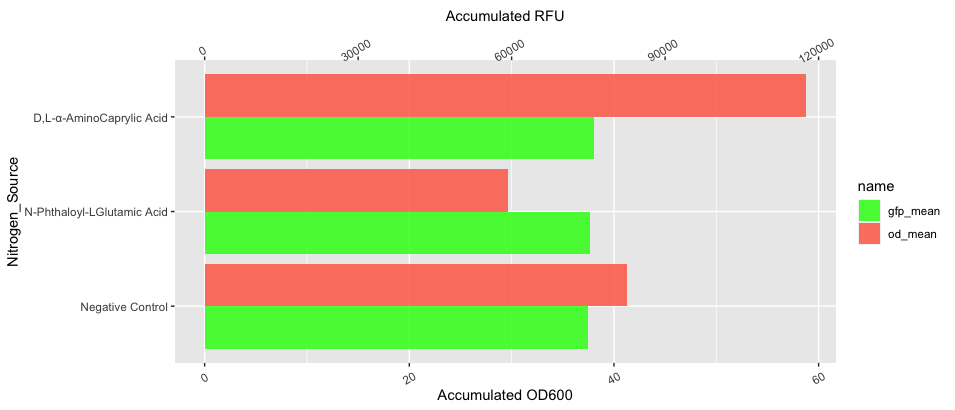
I'd also suggest swapping the axes, which you can do since the 2020 update to ggplot (v2.2 I think).
YOUR_DATA |>
dplyr:: mutate(val_adj = value / if_else(name == "gfp_mean", 2000, 1)) |>
ggplot( aes(x = val_adj, y = Nitrogen_Source, fill = name))
geom_col(position="dodge",alpha=0.8)
scale_x_continuous(
name="Accumulated OD600",
sec.axis = sec_axis(~.*2000,
name = "Accumulated RFU",))
scale_fill_manual(values = c("green", "tomato"))
theme(axis.text.x = element_text(angle = 30, vjust = 0.5))