This is a revised question that I posted earlier which has been improved for clarity.
I am attempting to make a scatterplot of CO2 rate data (y) at varying temperatures (t) with two categorical variables, depth (Mineral, Organic) and substrate (Calcareous, Metapelite, Peridotite) with fitted lines following the equation y = a*exp(b*t) where y = CO2 rate, a = basal respiration (intercept), b = slope and t = temperature (time equivalent). I have already fitted all of the exponential curves so I have the values for y, a, b and t for each data point. I am struggling to figure out how to plot the exponential curves using the function exp_funct <- function(y,a,b,t){y=a*exp(b*t)}
for each category of the two groups, depth and substrate. So far, I have produced the base scatterplot using GGplot2 colouring by depth and faceting by substrate but I do not know what the best approach is to fit the curves, I have attempted using geom_line and stat_function with no success.
Here's my attempt at some reproduceable code:
co2_data <- structure(list(chamber_temp = c(10, 10, 10, 10, 10, 15, 15, 15,
15, 15, 15, 19, 19, 19, 25, 25, 25, 25, 25, 25, 35, 35, 35, 35,
35, 35, 5, 5, 5, 5, 5, 5), substrate = c("Calcareous", "Metapelite",
"Metapelite", "Peridotite", "Peridotite", "Calcareous", "Calcareous",
"Metapelite", "Metapelite", "Peridotite", "Peridotite", "Calcareous",
"Calcareous", "Metapelite", "Calcareous", "Calcareous", "Metapelite",
"Metapelite", "Peridotite", "Peridotite", "Calcareous", "Calcareous",
"Metapelite", "Metapelite", "Peridotite", "Peridotite", "Calcareous",
"Calcareous", "Metapelite", "Metapelite", "Peridotite", "Peridotite"
), depth = c("Mineral", "Mineral", "Organic", "Mineral", "Organic",
"Mineral", "Organic", "Mineral", "Organic", "Mineral", "Organic",
"Mineral", "Organic", "Organic", "Mineral", "Organic", "Mineral",
"Organic", "Mineral", "Organic", "Mineral", "Organic", "Mineral",
"Organic", "Mineral", "Organic", "Mineral", "Organic", "Mineral",
"Organic", "Mineral", "Organic"), N.x = c(3, 6, 4, 5, 8, 6, 8,
7, 8, 8, 8, 3, 8, 4, 6, 8, 8, 8, 8, 8, 6, 8, 8, 8, 8, 8, 6, 8,
8, 8, 8, 8), basal_respiration = c(0.0092, 0.0124666666666667,
0.04935, 0.0101, 0.05785, 0.01315, 0.01415, 0.013, 0.0402, 0.01075,
0.05785, 0.0171, 0.01415, 0.03105, 0.01315, 0.01415, 0.013075,
0.0402, 0.01075, 0.05785, 0.01315, 0.01415, 0.013075, 0.0402,
0.01075, 0.05785, 0.01315, 0.01415, 0.013075, 0.0402, 0.01075,
0.05785), sd.x = c(0.00744781847254617, 0.00234065517893317,
0.00178978583448784, 0.00166132477258362, 0.0118691677407113,
0.00666175652512158, 0.00727284577825528, 0.00256059888828115,
0.00986798575481049, 0.00193833507349551, 0.0118691677407113,
0.00294448637286709, 0.00727284577825528, 0.000866025403784439,
0.00666175652512158, 0.00727284577825528, 0.00238012604708238,
0.00986798575481049, 0.00193833507349551, 0.0118691677407113,
0.00666175652512158, 0.00727284577825528, 0.00238012604708238,
0.00986798575481049, 0.00193833507349551, 0.0118691677407113,
0.00666175652512158, 0.00727284577825528, 0.00238012604708238,
0.00986798575481049, 0.00193833507349551, 0.0118691677407113),
se.x = c(0.0043, 0.000955568475364854, 0.000894892917243919,
0.000742967024840269, 0.0041963844982488, 0.00271965071286737,
0.00257133928416413, 0.000967815409397198, 0.00348885982193938,
0.000685304937340201, 0.0041963844982488, 0.0017, 0.00257133928416413,
0.00043301270189222, 0.00271965071286737, 0.00257133928416413,
0.000841501633985341, 0.00348885982193938, 0.000685304937340201,
0.0041963844982488, 0.00271965071286737, 0.00257133928416413,
0.000841501633985341, 0.00348885982193938, 0.000685304937340201,
0.0041963844982488, 0.00271965071286737, 0.00257133928416413,
0.000841501633985341, 0.00348885982193938, 0.000685304937340201,
0.0041963844982488), ci.x = c(0.0185014067379227, 0.00245636696547958,
0.00284794865810747, 0.00206280715944113, 0.00992287255356713,
0.00699108472177222, 0.00608025123040774, 0.00236815899497472,
0.00824984254536553, 0.00162048867457091, 0.00992287255356713,
0.00731450964057409, 0.00608025123040774, 0.00137803967327781,
0.00699108472177222, 0.00608025123040774, 0.00198983517147669,
0.00824984254536553, 0.00162048867457091, 0.00992287255356713,
0.00699108472177222, 0.00608025123040774, 0.00198983517147669,
0.00824984254536553, 0.00162048867457091, 0.00992287255356713,
0.00699108472177222, 0.00608025123040774, 0.00198983517147669,
0.00824984254536553, 0.00162048867457091, 0.00992287255356713
), N.y = c(3, 6, 4, 5, 8, 6, 8, 7, 8, 8, 8, 3, 8, 4, 6, 8,
8, 8, 8, 8, 6, 8, 8, 8, 8, 8, 6, 8, 8, 8, 8, 8), slope = c(0.120293333333333,
0.0593333333333333, 0.07685, 0.05602, 0.067475, 0.108913333333333,
0.15655, 0.0600714285714286, 0.08535, 0.057525, 0.067475,
0.0975333333333333, 0.15655, 0.09385, 0.108913333333333,
0.15655, 0.058125, 0.08535, 0.057525, 0.067475, 0.108913333333333,
0.15655, 0.058125, 0.08535, 0.057525, 0.067475, 0.108913333333333,
0.15655, 0.058125, 0.08535, 0.057525, 0.067475), sd.y = c(0.0326433842199814,
0.00744813175680094, 0.00456106712659804, 0.00374259268422306,
0.00379877799900366, 0.0244087448810189, 0.0131734581640509,
0.00707406396298344, 0.00967780966954816, 0.00357481268240529,
0.00379877799900366, 0.00594670777265315, 0.0131734581640509,
0.00225166604983954, 0.0244087448810189, 0.0131734581640509,
0.0085558250833653, 0.00967780966954816, 0.00357481268240529,
0.00379877799900366, 0.0244087448810189, 0.0131734581640509,
0.0085558250833653, 0.00967780966954816, 0.00357481268240529,
0.00379877799900366, 0.0244087448810189, 0.0131734581640509,
0.0085558250833653, 0.00967780966954816, 0.00357481268240529,
0.00379877799900366), se.y = c(0.0188466666666667, 0.00304068705686359,
0.00228053356329902, 0.00167373833080324, 0.00134307084165888,
0.00996482837004454, 0.00465752079973885, 0.0026737448578026,
0.00342162242218512, 0.00126388714460023, 0.00134307084165888,
0.00343333333333334, 0.00465752079973885, 0.00112583302491977,
0.00996482837004454, 0.00465752079973885, 0.00302494096754678,
0.00342162242218512, 0.00126388714460023, 0.00134307084165888,
0.00996482837004454, 0.00465752079973885, 0.00302494096754678,
0.00342162242218512, 0.00126388714460023, 0.00134307084165888,
0.00996482837004454, 0.00465752079973885, 0.00302494096754678,
0.00342162242218512, 0.00126388714460023, 0.00134307084165888
), ci.y = c(0.0810906617800115, 0.007816334916228, 0.00725767561259645,
0.00464704259594057, 0.00317585788379371, 0.0256154068032699,
0.0110132866353603, 0.0065424179794951, 0.00809085135929258,
0.00298861819339805, 0.00317585788379371, 0.0147724410388065,
0.0110132866353603, 0.0035829031505223, 0.0256154068032699,
0.0110132866353603, 0.00715284877149766, 0.00809085135929258,
0.00298861819339805, 0.00317585788379371, 0.0256154068032699,
0.0110132866353603, 0.00715284877149766, 0.00809085135929258,
0.00298861819339805, 0.00317585788379371, 0.0256154068032699,
0.0110132866353603, 0.00715284877149766, 0.00809085135929258,
0.00298861819339805, 0.00317585788379371), N = c(3, 6, 4,
5, 8, 6, 8, 7, 8, 8, 8, 3, 8, 4, 6, 8, 8, 8, 8, 8, 6, 8,
8, 8, 8, 8, 6, 8, 8, 8, 8, 8), co2_rate_u_m_h_g = c(0.0303333333333333,
0.0113333333333333, 0.0645, 0.0066, 0.129375, 0.0615, 0.1325,
0.0254285714285714, 0.132, 0.021, 0.14325, 0.085, 0.208,
0.17, 0.198666666666667, 0.71025, 0.0575, 0.344, 0.05225,
0.3115, 0.5155, 3.27125, 0.10375, 0.7835, 0.079, 0.6065,
-0.00766666666666667, 0.024625, 0.02675, 0.065125, 0.012125,
0.061), sd = c(0.0161658075373095, 0.0109483636524673, 0.0137719521734091,
0.00634822809924155, 0.0181102772716804, 0.0332009036021612,
0.0291498591028205, 0.00639940473422184, 0.0225895045161622,
0.0101136400116731, 0.0263425240140077, 0.00435889894354067,
0.0731358813637816, 0.0127279220613579, 0.0471197057149837,
0.302157834819581, 0.0214941852602047, 0.0326233921333407,
0.0116833214455479, 0.0554204706893452, 0.130053450550149,
1.288200932641, 0.0353138985184506, 0.129266060068814, 0.0200997512422418,
0.0639262968470052, 0.0164032517101539, 0.0136793640203045,
0.00948306761699881, 0.0180193348537461, 0.00918753036924038,
0.0156296421675518), se = c(0.00933333333333333, 0.00446965074449646,
0.00688597608670453, 0.00283901391331568, 0.0064029499339869,
0.0135542121374378, 0.0103060315211184, 0.00241874763794291,
0.00798659591351123, 0.00357571171736681, 0.00931348868193715,
0.00251661147842358, 0.0258574388301924, 0.00636396103067893,
0.0192365393053024, 0.106828926994785, 0.00759934207678533,
0.0115341109013965, 0.00413067791046458, 0.0195940953204931,
0.0530940988560248, 0.455447807500643, 0.012485348556265,
0.0457024538259631, 0.00710633520177595, 0.0226013589983308,
0.00669659946871877, 0.00483638553053828, 0.0033527707092152,
0.00637079693377748, 0.00324828251322361, 0.00552591298209755
), ci = c(0.0401580921443283, 0.0114896030154409, 0.0219142491554048,
0.00788236628321375, 0.0151405706956398, 0.0348422115168589,
0.0243698920725162, 0.00591846226021141, 0.0188852983847605,
0.00845521464359003, 0.0220229012042435, 0.0108281052473581,
0.0611431269419499, 0.0202529642690536, 0.0494490985187144,
0.252610271543504, 0.0179695885709161, 0.0272738383580029,
0.00976750116260314, 0.0463326729828588, 0.136482726098776,
1.07696293095078, 0.0295231579857332, 0.108069130674172,
0.0168038125580669, 0.0534437216064078, 0.0172141569548202,
0.0114362345155632, 0.00792804292904021, 0.0150645409315832,
0.0076809676067933, 0.0130667078496593)), row.names = c(NA,
-32L), class = "data.frame")
exp_funct <- function(y,a,b,t){y=a*exp(b*t)}
ggplot(co2_data ,
aes(x = chamber_temp, y = co2_rate_u_m_h_g, colour = substrate, linetype = substrate,
shape = substrate, fill = substrate),
ymax=co2_rate_u_m_h_g se, ymin=co2_rate_u_m_h_g-se)
geom_point(stat="identity", position = "dodge", width = 0.7)
geom_errorbar(aes(ymax=co2_rate_u_m_h_g se, ymin=co2_rate_u_m_h_g-se),
width=0.1, size=0.1, color="black")
facet_wrap(~depth)
stat_function(data = co2_data ,
aes(y= co2_rate_u_m_h_g, a = basal_respiration, b = slope, c = chamber_temp),
fun = exp_funct(y =co2_rate_u_m_h_g, a = basal_respiration, b = slope, t = chamber_temp))
Error in exp_funct(y = co2_rate_u_m_h_g, a = basal_respiration, b = slope, : object 'basal_respiration' not found In addition: Warning message: Ignoring unknown parameters: width
CodePudding user response:
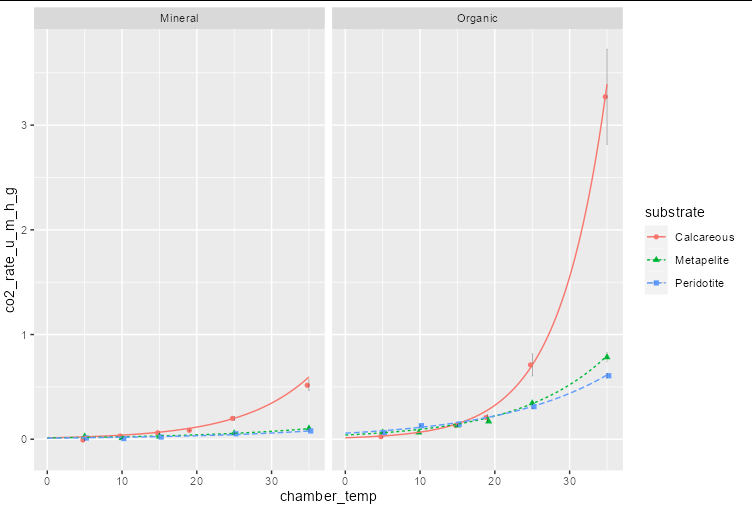
Additionally to Allan's answer, a collegue figured out how to do it using nls
library(tidyverse)
install.packages("devtools")
devtools::install_github("onofriAndreaPG/aomisc")
df <- expand.grid(substrate = c("Calcareous", "Metapelite", "Peridotite"),
depth = c("Mineral", "Organic"))
mods <- purrr::map(split(df, 1:nrow(df)),
function(x) nls(co2_rate_u_m_h_g ~ NLS.expoGrowth(chamber_temp, a, b),
data = dplyr::filter(co2_data, substrate == x[[1]], depth == x[[2]])))
pred_dfs <- purrr::map(split(df, 1:nrow(df)), function(x) expand.grid(substrate = x[[1]], depth = x[[2]], chamber_temp = seq(from = 0, to = 35)))
nls_pred <- function(x, y) {
pred <- predict(x, newdata = y)
y$pred <- pred
return(y)
}
preds <- purrr::map2_dfr(mods, pred_dfs, nls_pred)
ggplot(co2_data ,
aes(x = chamber_temp, y = co2_rate_u_m_h_g, linetype = substrate,
shape = substrate),
ymax=co2_rate_u_m_h_g se, ymin=co2_rate_u_m_h_g-se)
geom_line(data = preds, aes(y = pred, x = chamber_temp, colour = substrate), linewidth = 1)
geom_errorbar(aes(ymax=co2_rate_u_m_h_g se, ymin=co2_rate_u_m_h_g-se),
width=0.1, size=0.1, color="black")
geom_point(aes(fill = substrate), size = 3)
scale_shape_manual(values = c(21, 22, 24))
facet_wrap(~depth)
theme_bw()
theme(legend.position = "bottom")
CodePudding user response:
You can't feed parameters into stat_function that way. In this case, I would probably just generate a little summary data frame inside a geom_line call:
library(tidyverse)
ggplot(co2_data ,
aes(x = chamber_temp, y = co2_rate_u_m_h_g,
colour = substrate, linetype = substrate,
shape = substrate, fill = substrate,
ymax = co2_rate_u_m_h_g se, ymin = co2_rate_u_m_h_g - se))
geom_point(position = position_dodge(width = 0.7))
geom_errorbar(width = 0.1, size = 0.1, color = 'black')
facet_wrap(~depth)
geom_line(data = . %>% group_by(substrate, depth) %>%
summarize(chamber_temp = seq(0, 35, length.out = 100),
basal_respiration = mean(basal_respiration),
slope = mean(slope), se = mean(se),
co2_rate_u_m_h_g = basal_respiration *
exp(chamber_temp * slope)))