I am new to writing function in R, I have a dataset(created by me for practice from a large dataset), I want to loop through each column and flag the outlier.Any help or advice is appreciable! This is my dataset:
Time Temperature.C. Relative_Humidity
1 10/24/2022 16:45 32.2 50.0
2 10/24/2022 16:46 30.0 49.0
3 10/24/2022 16:47 31.0 50.0
4 10/24/2022 16:48 30.0 50.5
5 10/24/2022 16:49 30.0 50.0
6 10/24/2022 16:50 31.0 49.0
7 10/24/2022 16:51 32.2 51.0
8 10/24/2022 16:52 86.0 50.5
9 10/24/2022 16:53 30.0 50.0
10 10/24/2022 16:54 30.0 120.0
11 10/24/2022 16:55 30.0 50.0
12 10/24/2022 16:56 86.0 50.0
13 10/24/2022 16:57 30.0 51.0
14 10/24/2022 16:58 31.0 51.0
15 10/24/2022 16:59 31.0 50.0
16 10/24/2022 17:00 31.0 49.0
17 10/24/2022 17:01 3.0 52.0
18 10/24/2022 17:02 32.2 49.0
19 10/24/2022 17:03 30.0 2.0
structure(list(Time = c("10/24/2022 16:45", "10/24/2022 16:46",
"10/24/2022 16:47", "10/24/2022 16:48", "10/24/2022 16:49", "10/24/2022 16:50",
"10/24/2022 16:51", "10/24/2022 16:52", "10/24/2022 16:53", "10/24/2022 16:54",
"10/24/2022 16:55", "10/24/2022 16:56", "10/24/2022 16:57", "10/24/2022 16:58",
"10/24/2022 16:59", "10/24/2022 17:00", "10/24/2022 17:01", "10/24/2022 17:02",
"10/24/2022 17:03"), Temperature.C. = c(32.2, 30, 31, 30, 30,
31, 32.2, 86, 30, 30, 30, 86, 30, 31, 31, 31, 3, 32.2, 30), Relative_Humidity = c(50,
49, 50, 50.5, 50, 49, 51, 50.5, 50, 120, 50, 50, 51, 51, 50,
49, 52, 49, 2)), class = "data.frame", row.names = c(NA, -19L
))
I am expecting my output like this.
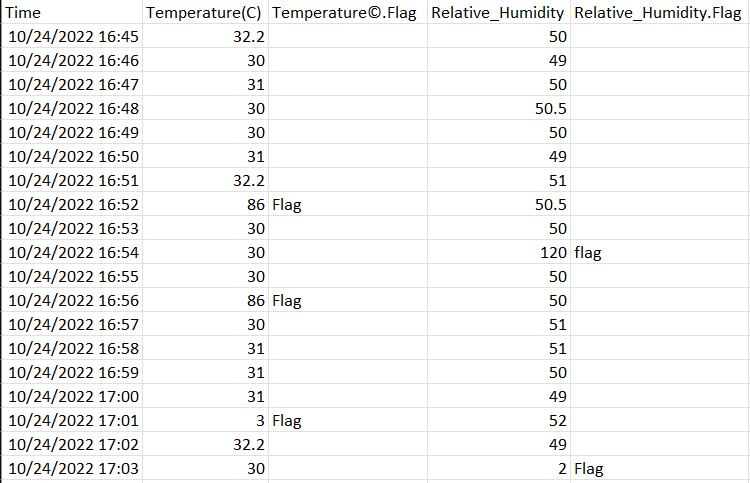
CodePudding user response:
Define your outlier limits See here:
You probably want to define an outlier as
- data point above Q3 IQR * 1.5
- data point under 5 percentile IQR * 1.5
These conditions fit best to your provided example:
library(dplyr)
df %>%
mutate(across(-Time, ~case_when(. > quantile(., probs = 0.75) IQR(.) * 1.5 ~ "FLAG",
. < quantile(., probs = 0.05) IQR(.) * 1.5 ~ "FLAG",
TRUE ~ ""), .names = "{col}_outlier")) %>%
relocate(Time, starts_with("Temperature"))
Time Temperature.C. Temperature.C._outlier Relative_Humidity Relative_Humidity_outlier
1 10/24/2022 16:45 32.2 50.0
2 10/24/2022 16:46 30.0 49.0
3 10/24/2022 16:47 31.0 50.0
4 10/24/2022 16:48 30.0 50.5
5 10/24/2022 16:49 30.0 50.0
6 10/24/2022 16:50 31.0 49.0
7 10/24/2022 16:51 32.2 51.0
8 10/24/2022 16:52 86.0 FLAG 50.5
9 10/24/2022 16:53 30.0 50.0
10 10/24/2022 16:54 30.0 120.0 FLAG
11 10/24/2022 16:55 30.0 50.0
12 10/24/2022 16:56 86.0 FLAG 50.0
13 10/24/2022 16:57 30.0 51.0
14 10/24/2022 16:58 31.0 51.0
15 10/24/2022 16:59 31.0 50.0
16 10/24/2022 17:00 31.0 49.0
17 10/24/2022 17:01 3.0 FLAG 52.0
18 10/24/2022 17:02 32.2 49.0
19 10/24/2022 17:03 30.0 2.0 FLAG
CodePudding user response:
This tidyverse example may be useful:
1. Create a sample dataset
library(tidyverse)
sample_data <- data.frame(
a = sample(1:10, 10, TRUE),
b = sample(1:10, 10, TRUE),
c = sample(1:10, 10, TRUE),
d = sample(1:10, 10, TRUE)
)
2. Detect which value(s) are outliers
Based on your description, it seems outliers are the maximum and minimum values, so we can use the range() function to get the range of each column.
outcome <- sample_data |>
mutate(across(everything(), ~ case_when( # note 1
.x == range(.x) ~ "Flag", # note 2
TRUE ~ ""
)))
note 1: You may need to replace everything() to c(var1, var2 ...) based on your dataset.
note 2: This part marks outliers as "Flag", and marks the rest as "".
3. Rename the outcome columns:
names(outcome) <- paste0(c("a", "b", "c", "d"), "_flag")
4. Combine data and outcome as one dataset
outcome <- bind_cols(sample_data, outcome)
5. Reorder the columns alphabetically
outcome <- outcome |>
select(order(colnames(outcome)))
The final outcome should look like below after you run outcome:
a a_flag b b_flag c c_flag d d_flag
1 10 1 Flag 10 7
2 5 9 Flag 5 8 Flag
3 1 Flag 8 6 5
4 7 3 7 8 Flag
5 3 7 8 4
6 4 2 7 4
7 6 5 7 5
8 2 4 5 5
9 1 Flag 2 2 Flag 5
10 10 Flag 5 10 Flag 1
Hope this example is helpful for your case.
